Abstract
Background
Ghrelin (GHRL) is a pivotal peptide regulator of food intake, energy balance, and body mass. Weight gain (WG) is a common side effect of the atypical antipsychotics (AAPs) used to treat schizophrenia (SZ). Ghrelin polymorphisms have been associated with pathogenic variations in plasma lipid concentrations, blood pressure, plasma glucose, and body mass index (BMI). However, it is unclear whether GHRL polymorphisms are associated with WG due to AAPs. Furthermore, there is no evidence of an association between GHRL polymorphisms and SZ or the therapeutic response to AAPs. We explored these potential associations by genotyping GHRL alleles in SZ patients and controls. We also examined the relation between these SNPs and changes in metabolic indices during AAP treatment in SZ subgroups distinguished by high or low therapeutic response.
Methods
Four SNPs (Leu72Met, -501A/C, -604 G/A, and -1062 G > C) were genotyped in 634 schizophrenia patients and 606 control subjects.
Results
There were no significant differences in allele frequencies, genotype distributions, or the distributions of two SNP haplotypes between SZ patients and healthy controls (P > 0.05). There was also no significant difference in symptom reduction between genotypes after 8 weeks of AAP treatment as measured by positive and negative symptom scale scores (PANSS). However, the -604 G/A polymorphism was associated with a greater BMI increase in response to AAP administration in both APP responders and non-responders as distinguished by PANSS score reduction (P < 0.001). There were also significant differences in WG when the responder group was further subdivided according to the specific AAP prescribed (P < 0.05).
Conclusions
These four GHRL gene SNPs were not associated with SZ in this Chinese Han population. The -604 G/A polymorphism was associated with significant BW and BMI increases during AAP treatment. Patients exhibiting higher WG showed greater improvements in positive and negative symptoms than patients exhibiting lower weight gain or weight loss.
Similar content being viewed by others
Background
Schizophrenia (SZ) is a severe brain disorder afflicting approximately 1% of the world's population and often leads to a lifetime of disability and emotional distress [1]. Family, twin, and adoption studies strongly indicate that genetics contribute to the etiology of SZ, probably by transmission of multiple susceptibility genes each exerting weak-to-moderate effects on predisposition [2, 3]. Many candidate susceptibility genes have been identified, including the dopamine receptor D2, neuregulin1, and disrupted in schizophrenia 1 (DISC-1) [4–6].
Epidemiological studies have also revealed that people with SZ are at greater risk for obesity, type 2 diabetes, dyslipidemia, and hypertension than the general population [7]. Recently, it was suggested that SZ patients are at increased risk of metabolic problems and that the associated symptoms are a serious threat to patient health [8]. Metabolic problems are often triggered by antipsychotic medication. Indeed, significant weight gain is common in AAP-treated SZ patients, especially patients administered clozapine, olanzapine, quetiapine, or risperidone [9].
The peptide ghrelin (product of the GHRL gene) is an important metabolic regulator produced by the stomach and pancreas. Specific SNPs of GHRL have been associated with variations in BMI, blood pressure, high-density lipoproteins, low-density lipoproteins, serum cholesterol, blood glucose, and metabolic syndrome [10–14]. Ghrelin, originally isolated from the rat stomach, stimulates food intake and controls energy balance [15, 16]. Studies on animal models revealed that GHRL increased food intake and adiposity [17, 18]. However, circulating GHRL levels were decreased in obese individuals, and serum GHRL levels were inversely correlated with BMI, suggesting that GHRL is not directly involved in most cases of obesity [19, 20]. Studies on the relationship between WG, circulating GHRL, and AAP have yielded inconsistent findings. Patients taking clozapine or olanzapine showed greater WG than patients on other antipsychotics [21]. In one study, plasma total GHRL and active GHRL were increased significantly immediately after olanzapine treatment, but the changes in BMI and body weight were not significant after 6 months of treatment [22]. In contrast, another study found that serum bioactive GHRL levels decreased significantly from baseline after 4 weeks of olanzapine monotherapy [23]. In humans, GHRL plays an important role in the long-term regulation of body weight (BW) as well as in the short-term regulation of appetite [19, 21]. Ghrelin stimulated preadipocyte differentiation, increased the BMI, and inhibited the anorexigenic effect of leptin [24]. Drug altering GHRL function may have distinct short- and long-term effects on BMI.
The human GHRL gene is located on chromosome 3 (3p25-p26), and consists of 4 exons and 3 introns [25, 26]. Several SNPs in the coding region of prepro-ghrelin have been described, but there is no known specific association between genetic variations in the human GHRL gene and SZ risk. However, region 3p25.1-26.1 is strongly associated with schizophrenia. In addition to GHRL, this region contains SYN2, HRH1, and GRM7, all candidate genes for schizophrenia. The positive symptoms of schizophrenia are associated with dysfunction in dopaminergic signaling, which is closely associated with a GHRL mutation [27–29]. Previous studies demonstrated that SNPs in GHRL were associated with high BMI; the Leu72Met allele was significantly associated with BMI and coronary artery disease [18, 19, 30], but this was not confirmed in other studies [26, 31, 32].
In light of the increase metabolic syndrome symptoms observed in SZ patients on AAPs, as well as the important role of GHRL as a metabolic regulator and the association between GHRL SNPs and metabolic indices, we hypothesized that (1) GHRL might be a candidate gene for SZ and that (2) allelic variants of GHRL might be associated with the propensity for BMI changes induced by AAP treatment. In addition, we tested (3) whether a putative relationship between GHRL SNPs and metabolic effects was specific to individual AAP types. Finally, (4) we examined if GHRL alleles influenced the clinical efficacy of AAPs. To these ends, we genotyped four SNPs and investigated whether they were associated with SZ and the therapeutic and metabolic effects of AAPs in the Han Chinese population.
Methods
The study group consisted of 634 diagnosed schizophrenic patients (332 males and 302 females; mean age: 27.14 ± 7.53 years). Patients were unrelated Han Chinese born and living in the North Henan province, and all their biological grandparents were of Han Chinese ancestry. Individuals with a history of severe medical complications, organic brain disease, any concomitant major psychiatric disorders, or substance dependence were excluded. All patients were recruited from the Department of Psychiatry of the Second Affiliated Hospital of Xinxiang Medical University, P.R. China. The consensus diagnoses were conducted by at least two experienced psychiatrists according to the Diagnostic and Statistical Manual of Mental Disorders, fourth Edition (DSM-IV) [33]. The patient group included paranoid (n = 309), catatonic (n = 50), collapse (n = 45), residual (67), and undifferentiated (n = 163) schizophrenic types.
The control group consisted of 606 healthy subjects (293 males and 313 females; mean age: 29.08 ± 7.80 years) recruited from communities and colleges within the same region and matched to the patient group for age, gender ratio, and Han ethnicity. Controls were recruited using a simple non-structured interview performed by psychiatrists. Individuals with personal or family histories of mental illness or neurological diseases were excluded. The objectives and procedures of the study were explained to all subjects and written informed consent was obtained. The Ethical Committee of the Department of Psychiatry of the Second Affiliated Hospital of Xinxiang Medical University approved this study.
Three hundred and eighty patients were evaluated using the Positive and Negative Symptom Scale (PANSS) [34] before and after an 8-week administration of antipsychotic medications. Only those patients with total PANSS scores ≥ 60 before treatment were included. The reduction in PANSS scores from baseline after the 8-week treatment regime was used to evaluate the efficacy of each AAP. Patients were divided into 2 groups based on the reduction in PANSS score, a responder group exhibiting a > 50% reduction and none-responder group exhibiting a ≤ 50% reduction [35].
We excluded patients with incomplete clinical data. A total of 569 patients were treated by monotherapy using an AAP not previously prescribed. Patients were treated with clozapine (n = 103, 100-700 mg/d), risperidone (n = 181, 2-6 mg/d), olanzapine (n = 60, 5-20 mg/d), quetiapine (n = 126, 400-750 mg/d), ziprasidone (n = 61, 80-160 mg/d), or aripiprazole (n = 38, 10-30 mg/d). Body weight and BMI was measured before and after 4 weeks of AAP treatment. Individual BMIs were calculated as BMI = weight (kg)/height2 (m).
Peripheral blood samples were obtained from the subjects and genomic DNA was prepared using the QIAamp DNA blood Mini Kit (QIAGEN, Hilden, Germany). Four SNPs [rs696217 (Leu72Met), rs26802 (-501A/C), rs27647 (-604 G/A), and rs26311 (-1062 G > C)] were selected according to the dbSNP database http://www.ncbi.nlm.nih.gov/SNP/. The SNPs rs27647 and rs26311 are located in the promoter region, rs26802 in intron 1, and rs696217 in exon 3 of GHRL. All these SNPs effect GHRL function and have been linked to metabolic symptoms. The rs696217 amino-acid change (Leu72Met) affects the tail of the pro-ghrelin molecule, but it is not known how this affects GHRL expression or activity (Table 1). The four SNPs were detected by polymerase chain reaction (PCR)-based restriction fragment length polymorphism (PCR-RFLP) analysis.
The primers of the four SNPs investigated are shown in Table 1. The conditions used for PCR amplification included an initial denaturation step at 94°C for 5 min, followed by 36 cycles of 94°C for 30 s, 55-65°C for 30 s, and 72°C for 1 min, followed by a final extension at 72°C for 10 min. Small volumes (10 μl) of these PCR products were completely digested with 2U of restriction enzyme (Dra I for -604 G/A, Mwo I for -501A/C, Bsr I for Leu72Met, and Bcn I for -1062 G > C). The fragments were separated on 2-4% agarose gels and visualized under ultraviolet light after staining with ethidium bromide.
The statistical power of the sample size was calculated by the genetic power calculator (GPC, http://pngu.mgh.harvard.edu/~purcell/gpc/cc2.html) [36]. Deviations in the genotype counts from Hardy-Weinberg equilibrium were tested using a χ2 goodness-of-fit test. Statistical differences in genotypic, allelic, and haplotypic distributions between SZ and control subjects were evaluated by the χ2 test with a significance level of 0.05. Odds ratios (OR) and 95% confidence intervals (95% CI) were calculated to evaluate the effects of different alleles on SZ risk. Pair-wise linkage disequilibrium (LD) analysis was applied to detect the inter-marker relationship using D' and r2 values. Case-control association analysis was performed by SHEsis software http://analysis.bio-x.cn/myAnalysis.php[37], a powerful software platform for analyses of LD, haplotype construction, and genetic association at polymorphic loci. Associations between response to a specific AAP and genotype were determined by t-tests and analysis of variance (ANOVA) tests using SPSS 13.0 software. Results were considered significant at P < 0.05 (two-tailed).
The size of our sample was sufficient to detect a significant difference with a power of more than 70% assuming an OR value for AA of 1.5 with a minor allele frequency of 0.1 and type I error rate set at 0.05.
Results
Four SNPs in the GHRL locus were analyzed: Leu72Met, -501A/C, -604 G/A, and -1062 G > C. As shown in Table 2, none of the genotype distributions of these four SNPs showed significant deviation from Hardy-Weinberg equilibrium, and none of the allele frequencies or the genotype distribution differed between patients and controls (P > 0.05). There was also no significant association between any allele or genotype and SZ when patients were subdivided by gender (Table 3).
To further analyze the haplotype structure in our sample, pair-wise linkage disequilibrium (LD) of the four SNPs in the control group was computed using the standardized measures D' and r2 values. There was strong LD in Leu72Met and -501A/C, so haplotype analyses were performed (Table 4). However, the haplotypes constructed from two SNPs showed no significant differences between patients and controls (Table 5).
Of the 634 patients with SZ, 380 completed the PANSS to assess psychopathological syndromes. The results revealed that there were no significant differences in PANSS score reduction among the different genotypes of the four SNPs (-604 G/A, -501A/C, Leu72Met and -1062 G > C) after 8 weeks treatment with AAPs (Table 6). However, in the responder group with the larger decreases in PANSS scores (> 50%), there was a significant association between BW and BMI increase (Table 7). The responder group exhibited significantly greater BW and BMI increases than none responders, and patients in specific AAP treatment groups with high weight gain showed greater improvements than those with low weight gain when subdivided according to drug (Table 8).
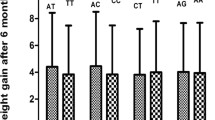
The main clinical and biochemical characteristics of the schizophrenic patients were analyzed with nonparametric tests. There was a significant association between BW and BMI measured before and after 4-week AAP treatment (P = 0.005 and 0.004 respectively). Patients with the -604 G/A exhibited significantly higher BWs and BMIs after treatment (P = 0.028 and 0.011, respectively) (Table 9). Similarly, paranoid SZ patients (n = 309) demonstrated greater WG and BMI increases (P = 0.020 and 0.011, respectively). In addition, there were significant differences in the BW and BMI increases between G allele carriers and homozygous allele A carriers in patients harboring SNP-604 G/A (P = 0.039 and 0.013, respectively).
Discussion
The associations between GHRL polymorphisms and SZ risk, changes in weight/BMI, and therapeutic responses to AAPs were evaluated in a population of SZ patients of Han Chinese ethnicity. While we found no association between GHRL gene polymorphisms and SZ susceptibility in this case-control study, analysis did reveal significant BW and BMI increases during AAP treatment in patients harboring the -604 G/A polymorphism.
To our knowledge, no previous study has examined the association between GHRL gene polymorphisms and susceptibility to SZ. Our study revealed no significant differences in allele and genotype frequency of four GHRL SNPs between schizophrenic patients and controls even when patients were subdivided by gender. Thus, GHRL is not a likely SZ risk gene despite the fact that it is in a susceptibility locus (3p25-p26). Furthermore, we also examined paranoid SZ cases in light of the study by Scassellati et al. [38]. Again, we found no significant differences in the frequency of these four SNPs or the genotype distribution between paranoid patients and controls, but this could reflect the relatively small sample size of paranoid schizophrenic patients in our cohort. Furthermore, we found no haplotypes with significantly higher frequency between cases and controls. Therefore, we suggest that GHRL is not a predisposing gene for SZ in the Chinese Han population.
In the present study, no association between PANSS reduction during AAP treatment and GHRL gene polymorphisms was found. However, the magnitude of the PANSS score reduction was significantly associated with the increase in BW and BMI during AAP treatment. Meanwhile, the reduction rate of PANSS total score in responder and none-responder groups had significant association with BW and BMI increase. The same finding was also revealed when patients were subdivided according the specific AAP taken. Atypical antipsychotics induced weight gain in a significant fraction of SZ patients [9], but factors that are predictive of weight gain during AAP therapy are unclear. We found that patients exhibiting the greatest weight gains while receiving olanzapine, risperidone, clozapine, or quetiapine also showed greater improvements in symptoms than those showing lower weight gain. This result is in partial accord with a previous study that found olanzapine-induced weight gain correlated negatively with baseline BMI and positively with clinical global improvement and the length of olanzapine treatment [39].
The GHRL gene may be a promising candidate underlying AP-induced weight gain [40]. We found significant differences between the three -604 G/A genotypes, with patients harboring AA showing the greatest weight gain and increase in BMI. In addition to BMI, -604 G/A has been linked to variations in blood pressure [12]. Previous studies have also reported that the Leu72Met polymorphism was significantly associated with BMI [10, 11]. However, we found no association between Leu72Met polymorphisms and the AAP-induced BMI increase, consistent with previous findings [26, 31, 32]. In addition to the significant association between the AA genotype and BMI, we also found that paranoid SZ patients demonstrated higher weight gain than patients with other subtypes of SZ, including catatonic, collapse, residual, and undifferentiated patients. Therefore, our results provide suggestive evidence for a link between -604 G/A and metabolic syndrome in paranoid SZ.
Conclusion
While we did not find an association between GHRL alleles and susceptibility to SZ in the Chinese Han population, the -604 G/A polymorphism, and particularly the AA genotype, was associated with larger increases in BW and BMI in SZ patients under treatment with AAPs. Surprisingly, patients showing the greatest weight gain also showed the greatest improvements in symptoms. In order to more precisely define the impact of antipsychotic medications on metabolic parameters, control of patient subtype, sample size, as well as monitoring of multiple metabolic indices during antipsychotic therapy are of paramount importance.
Abbreviations
- SZ:
-
Schizophrenia
- GHRL :
-
Ghrelin
- WG:
-
Weight gain
- AAPs:
-
Atypical antipsychotics
- BMI:
-
Body mass index
- SNPs:
-
Single nucleotide polymorphisms
- BW:
-
Body weight
- DSM-IV:
-
Diagnostic and statistical manual of mental disorders fourth edition
- PCR-RFLP:
-
Polymerase chain reaction-based-restriction fragment length polymorphism
- OR:
-
Odds ratio
- 95% CI:
-
95% confidence intervals
- LD:
-
Linkage disequilibrium
- ANOVA:
-
Analysis of variance
- PANSS:
-
Positive and negative symptom scale.
References
Lewis DA, Lieberman JA: Catching up on schizophrenia: natural history and neurobiology. Neuron. 2000, 28 (2): 325-334. 10.1016/S0896-6273(00)00111-2.
Hallmayer J: The epidemiology of the genetic liability for schizophrenia. Aust N Z J Psychiatr. 2000, 34 (Suppl): 47-55. 10.1046/j.1440-1614.2000.00769.x.
McGuffin P, Owen MJ, Farmer AE: Genetic basis of schizophrenia. Lancet. 1995, 346 (8976): 678-682. 10.1016/S0140-6736(95)92285-7.
Talkowski ME, Bamne M, Mansour H, Nimgaonkar VL: Dopamin genes and schizophrenia: case closed or evidence pending?. Schizophr Bull. 2007, 33 (5): 1071-1081. 10.1093/schbul/sbm076.
Tomppo L, Hennah W, Miettunen J, Järvelin MR, Veijola J, Ripatti S: Association of variants in DISC1 with psychosis-related traits in a large population cohort. Arch Gen Psychiatry. 2009, 66 (2): 134-141. 10.1001/archgenpsychiatry.2008.524.
Zhang HX, Zhao JP, Lv LX, Li WQ, Xu L, Ouyang X: Explorative study on the expression of neuregulin-1 gene in peripheral blood of schizophrenia. Neurosci Lett. 2008, 438 (1): 1-5. 10.1016/j.neulet.2007.09.051.
Citrome L, Vreeland B: Schizophrenia, obesity, and antipsychotic medications: what can we do?. Postgrad Med. 2008, 120 (2): 18-33. 10.3810/pgm.2008.07.1786.
Herberth M, Koethe D, Cheng TM, Krzyszton ND, Schoeffmann S, Guest PC: Impaired glycolytic response in peripheral blood mononuclear cells of first-onset antipsychotic-naive schizophrenia patients. Mol Psychiatry. 2011, 16 (8): 848-859. 10.1038/mp.2010.71.
Rummel-Kluge C, Komossa K, Schwarz S, Hunger H, Schmid F, Lobos CA: Head-to-head comparisons of metabolic side effects of second generation antipsychotics in the treatment of schizophrenia: a systematic review and meta-analysis. Schizophr Res. 2010, 123 (2-3): 225-233. 10.1016/j.schres.2010.07.012.
Ando T, Ichimaru Y, Konjiki F, Shoji M, Komaki G: Variations in the preproghrelin gene correlate with higher body mass index, fat mass, and body dissatisfaction in young Japanese women. Am J Clin Nutr. 2007, 86 (1): 25-32.
Kuzuya M, Ando F, Iguchi A, Shimokata H: Preproghrelin Leu72Met variant contributes to overweight in middle-aged men of a Japanese large cohort. Int J Obes (Lond). 2006, 30 (11): 1609-1614. 10.1038/sj.ijo.0803296.
Mager U, Kolehmainen M, Lindström J, Eriksson JG, Valle TT, Hämäläinen H: Association between ghrelin gene variations and blood pressure in subjects with impaired glucose tolerance. Am J Hypertens. 2006, 19 (9): 920-926. 10.1016/j.amjhyper.2006.02.017.
Choi HJ, Cho YM, Moon MK, Choi HH, Shin HD, Jang HC: Polymorphisms in the ghrelin gene are associated with serum high-density lipoprotein cholesterol level and not with type 2 diabetes mellitus in Koreans. J Clin Endocrinol Metab. 2006, 91 (11): 4657-4663. 10.1210/jc.2005-2549.
Steinle NI, Pollin TI, O'Connell JR, Mitchell BD, Shuldiner AR: Variants in the ghrelin gene are associated with metabolic syndrome in the Old Order Amish. J Clin Endocrinol Metab. 2005, 90 (12): 6672-6677. 10.1210/jc.2005-0549.
Kojima M, Hosoda H, Date Y, Nakazato M, Matsuo H, Kangawa K: Ghrelin is a growth-hormone-releasing acylated peptide from stomach. Nature. 1999, 402 (6762): 656-660. 10.1038/45230.
Wren AM, Seal LJ, Cohen MA, Brynes AE, Frost GS, Murphy KG, Bloom SR: Ghrelin enhances appetite and increases food intake in humans. J Clin Endocrinol Metab. 2001, 86 (12): 5992-10.1210/jc.86.12.5992.
Tschöp M, Smiley DL, Heiman ML: Ghrelin induces adiposity in rodents. Nature. 2000, 407 (6806): 908-913. 10.1038/35038090.
Wren AM, Small CJ, Ward HL, Murphy KG, Dakin CL, Taheri S: The novel hypothalamic peptide ghrelin stimulates food intake and growth hormone secretion. Endocrinology. 2000, 141 (11): 4325-4328. 10.1210/en.141.11.4325.
Shiiya T, Nakazato M, Mizuta M, Date Y, Mondal MS, Tanaka M: Plasma ghrelin levels in lean and obese humans and the effect of glucose on ghrelin secretion. J Clin Endocrinol Metab. 2002, 87 (1): 240-244. 10.1210/jc.87.1.240.
Yildiz BO, Suchard MA, Wong ML, McCann SM, Licinio J: Alterations in the dynamics of circulating ghrelin, adiponectin, and leptin in human obesity. Proc Natl Acad Sci USA. 2004, 101 (28): 10434-10439. 10.1073/pnas.0403465101.
Himmerich H, Fulda S, Künzel HE, Pfennig A, Dzaja A, Cummings DE: Ghrelin plasma levels during psychopharmacological treatment. Neuropsychobiology. 2005, 52: 11-16. 10.1159/000086171.
Murashita M, Kusumi I, Inoue T, Takahashi Y, Hosoda H, Kangawa K: Olanzapine increases plasma ghrelin level in patients with schizophrenia. Psychoneuroendocrinology. 2005, 30: 106-110. 10.1016/j.psyneuen.2004.05.008.
Hosojima H, Togo T, Odawara T, Hasegawa K, Miura S, Kato Y: Early effects of olanzapine on serum levels of ghrelin, adiponectin and leptin in patients with schizophrenia. J Psychopharmacol. 2006, 20: 75-79.
Korbonits M, Goldstone AP, Gueorguiev M, Grossman AB: Ghrelin-a hormone with multiple functions. Front Neuroendocrinol. 2004, 25 (1): 27-68. 10.1016/j.yfrne.2004.03.002.
Wajanrajch MP, Ten IS, Gertner JM, Leibel RL: Genomic organization of human ghrelin gene. J Endocr Genet. 2000, 1: 231-233.
Garcia EA, King P, Sidhu K, Ohgusu H, Walley A, Lecoeur C: The role of ghrelin and ghrelin-receptor gene variants and promoter activity in type 2 diabetes. Eur J Endocrinol. 2009, 161 (2): 307-315. 10.1530/EJE-09-0122.
Correll CU, Malhotra AK: Pharmacogenetics of antipsychotic-induced weight gain. Psychopharmacology (Berl). 2004, 174 (4): 477-489.
Horvath TL, Castañeda T, Tang-Christensen M: Ghrelin as a potential anti-obesity target. Curr Pharm Des. 2003, 9 (17): 1383-1395. 10.2174/1381612033454748.
Online datebase center, sponsored by NARSAD and the Mental Health Research Association [DB/OL]. 2005, http://www.schizophreniaforum.org/res/sczgene/default.asp
Zhang Q, Huang WD, Lv XY, Yang YM: The association of ghrelin polymorphisms with coronary artery disease and ischemic chronic heart failure in an elderly Chinese population. Clin Biochem. 2011, 44 (5-6): 386-390. 10.1016/j.clinbiochem.2010.12.013.
Hubácek JA, Bohuslavová R, Skodová Z, Adámková V: Variants within the ghrelin gene-association with HDL-cholesterol, but not with body mass index. Folia Biol (Praha). 2007, 53 (6): 202-206.
Mager U, Lindi V, Lindström J, Eriksson JG, Valle TT, Hämäläinen H: Association of the Leu72Met polymorphism of the ghrelin gene with the risk of type 2 diabetes in subjects with impaired glucose tolerance in the Finnish Diabetes Prevention Study. Diabet Med. 2006, 23 (6): 685-689. 10.1111/j.1464-5491.2006.01870.x.
American Psychiatric Association: Diagnostic and Statistical Manual of Mental Disorders. 1994, Washington, DC: American Psychiatric Press, fourth
Kay SR, Opler LA, Lindenmayer JP: Reliability and validity of the positive and negative syndrome scale for schizophrenics. Psychiatry Res. 1988, 23 (1): 99-110. 10.1016/0165-1781(88)90038-8.
Zhang ML: Relationship in dosage, serum concentration and clinical response of olanzap ine in schizophrenic patients. Chin J Psychiatry. 2003, 36 (3): 139-
Purcell S, Cherny SS, Sham PC: Genetic Power Calculator: design of linkage and association genetic mapping studies of complex traits. Bioinformatics. 2003, 19 (1): 149-150. 10.1093/bioinformatics/19.1.149.
Shi YY, He L: SHEsis, a powerful software platform for analyses of linkage disequilibrium, haplotype construction, and genetic association at polymorphism loci. Cell Res. 2005, 15 (2): 97-98. 10.1038/sj.cr.7290272.
Scassellati C, Bonvicini C, Perez J, Bocchio-Chiavetto L, Tura GB, Rossi G: Association study of -1727 A/T, -50 C/T and (CAA)n repeat GSK-3beta gene polymorphisms with schizophrenia. Neuropsychobiology. 2004, 50 (1): 16-20. 10.1159/000077936.
Ujike H, Nomura A, Morita Y, Morio A, Okahisa Y, Kotaka T: Multiple genetic factors in olanzapine-induced weight gain in schizophrenia patients: a cohort study. J Clin Psychiatry. 2008, 69: 1416-1422. 10.4088/JCP.v69n0909.
Correll CU, Malhotra AK: Pharmacogenetics of antipsychotic-induced weight gain. Psychopharmacology. 2004, 174 (4): 477-489.
Acknowledgements
We thank Zhaoxi ZHONG, Zheng ZHAO, Jun CHENG, Yunhong DU, Yuchun LI, Yuling LI, Dexiang DUAN and Fang GUO(Department of Psychiatry of the Second Affiliated Hospital of Xinxiang Medical University, Xinxiang, 453002, PR China) for their works of collect the clinical materiales, and thank Yan RUAN, Lifang WANG, Lei WANG, Tianlan LU, Jian QIN, Zhilin LUAN, Lin TIAN and Hao YAN. (Institute of Mental Health, Peking University, Beijing, 100083, PR China) for their assistance in doing experiment.
The research was supported by Ministry of Health Research Fund of the People's Republic of China (Grant No.200801009), and the Supported by Program for Innovative Research Team (in Science and Technology) in University of Henan Province (Grant No.2008IRTSTHN008), the National Natural Science Foundation of China (30530290, 81071090, 81071091), the National High Technology Research and Development Program of China (2009AA022702), the National Basic Research Program of China (2007CB512301).
Author information
Authors and Affiliations
Corresponding authors
Additional information
Competing interests
The authors declare that they have no competing interests.
Authors' contributions
LL, WY and DZ participated in the design of the study and made final approval of the version to be published. YY were involved in drafting the manuscript and data analysis. WL, YY, HY, HZ, XS, BX, and GY carried out the molecular genetic examination. YY, WY, CJ, and JZ conducted sample selection and data management. All authors read and approved the final manuscript.
Yongfeng Yang, Wenqiang Li contributed equally to this work.
Authors’ original submitted files for images
Below are the links to the authors’ original submitted files for images.
Rights and permissions
Open Access This article is published under license to BioMed Central Ltd. This is an Open Access article is distributed under the terms of the Creative Commons Attribution License ( https://creativecommons.org/licenses/by/2.0 ), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
About this article
Cite this article
Yang, Y., Li, W., Zhao, J. et al. Association between Ghrelin gene (GHRL) polymorphisms and clinical response to atypical antipsychotic drugs in Han Chinese schizophrenia patients. Behav Brain Funct 8, 11 (2012). https://doi.org/10.1186/1744-9081-8-11
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/1744-9081-8-11