Abstract
Background
A mycovirus previously identified in brown discolored fruiting bodies of the cultivated mushroom Flammulina velutipes was characterized. We tentatively named the virus the F. velutipes browning virus (FvBV).
Results
Purified FvBV particles contained two dsRNA genomes (dsRNA1 and 2). The complete sequence of dsRNA1 was 1,915 bp long, containing a single open reading frame (ORF) that encoded 580 amino acids of a putative 66-kDa RNA-dependent RNA polymerase (RdRp). dsRNA2 was 1,730 bp long containing a single ORF encoding 541 amino acids of a putative 60-kDa coat protein (CP1). Phylogenetic analysis of the RdRp sequences revealed FvBV to be a Partitivirus, most closely related to Chondrostereum purpureum cryptic virus. An RT-PCR assay was developed for the amplification of a 495-bp cDNA fragment from dsRNA encoding the CP1. When wild F. velutipes isolated from various parts of Japan were examined by RT-PCR assay, three isolates from the central region of Japan contained FvBV. One wild strain infected with FvBV was isolated in Nagano prefecture, where brown discoloration of white cultivated strains has occurred. Fruiting bodies produced by virus-harboring and virus-free F. velutipes were compared.
Conclusions
Cap color of the fruiting bodies of F. velutipes that contained Partitivirus FvBV was darker than FvBV-free fruiting bodies. The use of RT-PCR enabled association of FvBV and dark brown color of the fruiting body produced by F. velutipes strains.
Similar content being viewed by others
Background
At the time mycoviruses were discovered in the white button mushroom, Agaricus bisporus, in 1962 [1], Lentinula edodes was the only artificially cultivated mushroom in Japan. Since then, the number of cultivated mushroom species has greatly increased: L. edodes, Flammulina velutipes, Hypsizygus marmoreus, Pholiota nameko, Grifola frondosa, and Pleurotus eryngii are cultivated daily. As the mushroom industry in Japan continues to grow, various abnormal symptoms during cultivation have been observed. Symptom that resembled abiotic disorders of A. bisporus[2] first became apparent with cultivated strains of F. velutipes[3, 4]. One of the abnormal symptoms was the spontaneous appearance of brown discolored fruiting bodies among the white ones. We detected spherical virus particles in the brown discolored fruiting bodies of two different cultivated F. velutipes[3, 5]. We tentatively named the virus FvBV, for Flammulina velutipes browning virus.
Browning is also a typical symptom observed in MVX (Mushroom virus X) disease in A. bisporus[6–8]. With MVX, complex patterns of more than 26 dsRNA bands are detected by agarose gel electrophoresis. Four low molecular weight dsRNAs have been specifically linked with the browning syndrome [6] together with some bacterial agent [8].
In the present study, we determined the sequences of the dsRNA genomes of FvBV. The two dsRNA isolated from the purified virus particles potentially coded for RNA-dependent RNA polymerase (RdRp) and a coat protein. Based on the phylogenetic analysis of the RdRp sequence, FvBV belongs to the family Partitiviridae. From the nucleotide sequence of the FvBV coat protein, RT-PCR primers were designed. To estimate the influence of FvBV on fruiting body color, fruiting bodies produced by virus-harboring strains and virus-free strains of F. velutipes were compared. Based on the RT-PCR assay developed in this study, FvBV and the dark brown cap color of the fruiting bodies were clearly linked.
Results
Molecular characterization of FvBV
The complete nucleotide sequences of the two dsRNAs isolated from the purified virions (Figure 1) were determined. The sequences are deposited in DDBJ under the accession numbers of AB465308 and AB465309.
dsRNA1: The sequence of dsRNA1 was 1,915 bp in length, containing a single open reading frame (ORF) starting at position nt 56 and ending at nt 1798, which encoded a putative protein of 580 amino acids with a deduced molecular mass of 66,716 Da (Figure 2). A PSI-BLAST [9] search of the deduced amino acid sequence of dsRNA1, revealed the presence of the conserved motif (pFam cd01699 RNA_dep_RNAP, 3e-6)) [10] for RdRp within the ORF. RdRp is an essential protein encoded in the genomes of all RNA containing viruses with no DNA stage.
Sequence identities and E values of proteins deposited in GenBank that showed significant similarity with the putative RdRp encoded by dsRNA 1 are listed in Table 1. All of these viruses belonged to the family Partitiviridae[11]. Multiple alignments of RdRp amino acid sequences of the viruses listed in Table 1 were constructed using the MAFFT program [12]. Conserved motifs III-VIII of RdRp of dsRNA viruses [13] were found (Figure 3). An unrooted phylogenetic tree based on the alignment with bootstrap of 1000 was made (Figure 4). RdRp of FvBV was grouped together with a partitivirus in the basidiomycete fungus Chondrostereum purpureum and placed adjacent to a group containing two partitiviruses of pathogenic basidiomycete fungus: Helicobasidium mompa dsRNA mycovirus and Heterobasidion RNA Virus (Figure 4).
Alignment of RdRp sequences. The six conserved motifs (III~VIII) of RdRp are located. Identical residues are in black background. : and . indicate similar residues. FvBV: F. velutipes browning virus, CpCV1: C. purpureum cryptic virus1, HmV: H. mompa dsRNA mycovirus, HetRV: Heterobasidion RNA Virus, VfV1: Vicia faba partitivirus 1, OMVII: Oyster mushroom isometric virus II, FvIV: F. velutipes isometic virus, ScV: Sclerotinia sclerotiorum partitivirus S
Unrooted phylogenetic tree generated from amino acid sequences of RdRp. The unrooted phylogenetic tree of the aligned sequences was constructed based on the neighbor-joining method with a 1,000 replicate bootstrap search. The alignment was carried out with the program L-INS-i of MAFFT and visualized on the Web server http://align.genome.jp/mafft/. Numbers at the nod indicate percentage of bootstrap supports from 1,000 replicates.
dsRNA2: The sequence was 1,730 bp, with a single ORF starting at nt 71 and ending at 1,460 (Figure 2). The deduced amino acid sequence coded for putative protein of 541 amino acids with a molecular mass of 60,513 Da. Sequence identities and E values of proteins deposited in GenBank that showed significant similarity with the putative coat protein encoded by dsRNA 2 are listed in Table 2. Multiple alignments of coat protein amino acid sequences of the viruses listed in Table 2 are shown (Figure 5).
Fruiting body production and RT-PCR analysis
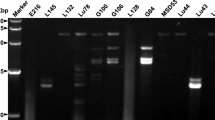
Among the 14 wild isolates (Table 3) examined for dsRNA, five from the central region of Japan contained dsRNA (Figure 6). Based on the RT-PCR analysis, three of the five isolates contained FvBV (Figure 7). Among the five dsRNA-harboring wild isolates, isolate No. 4 did not produce fruiting bodies. Also, Isolate No. 6 did not produce enough fruiting bodies (yield 37 g) to make an accurate comparison that it was eliminated from the test. Other dsRNA-harboring wild isolates, including isolate 5 (yield 107 g), 7 (yield 62 g) and 8 (yield 100 g) produced enough fruiting bodies for color comparison. As shown in the photograph (Figure 8), dsRNA-free wild isolate No. 2 (yield 104 g) and No. 11 (yield 98 g) produced fruiting bodies with paler-colored caps while fruiting bodies of the two isolates (No.7 and 8) containing FvBV were dark brown (Figure 8). Color of fruiting bodies produced by isolate No.5, which contained dsRNA but not FvBV, was much paler compared to isolate 7 and 8. All the F. velutipes harboring FvBV (two wild and one cultivated) produced dark brown fruiting bodies.
Fruiting body production of F. velutipes. From left to right; Fruiting bodies of virus-free (isolate No. 11, 2 and S6) and virus-containing (isolate No. 8, 5, 7 and S6B) F. velutipes. The result shows that all of the F. velutipes containing FvBV (Isolate No.7, 8 and S6B) produced fruiting bodies with dark brown caps.
Discussion
In our previous study, we investigated the presence of dsRNA elements in cultivated strains of F. velutipes to determine whether viruses are involved in the malformation of fruiting bodies. All of these strains were, however, dsRNA free [3]. Unexpectedly, spherical mycoviruses were detected in F. velutipes that showed brown discoloration during cultivation. This finding suggested that mycovirus causes brown discoloration of the mushroom. We tentatively named the virus Flammulina velutipes browning virus, or FvBV. Because browning reduces the crop value of white mushrooms, FvBV was further characterized in this study.
In this study, the sequences of two dsRNAs contained in the viral particles of FvBV were determined. dsRNA1 coded for putative RdRp with a sequence showing significant homology with members of the family Partitiviridae (Table 1). The family Partitiviridae consists of fungal and plant viruses with isometric virions, 30-35 nm in diameter, with a genome of two dsRNA segments each of 1.4-2.2 kb. The viruses are associated with latent infections of fungal or plant hosts [11]. dsRNA2 coded for putative coat protein with a sequence showing high homology with partitiviruses (Table 2). But the E-values were much larger compared to RdRp sequences.
It is noted that one of the three wild isolates (Isolate No. 8) harboring FvBV was isolated in Nagano prefecture, where spontaneous discoloration of cultivated F. velutipes has occurred at two independent locations (Figure 6). Because the mycelium of strain S6B does not show incompatibility against strain S6 (data not shown), it is unlikely that strain S6B is a wild F. velutipes contaminated during cultivation. Thus, presumably, there was horizontal transfer of the virus from a wild isolate to a cultivated strain of F. velutipes within Nagano prefecture.
The result of mushroom cultivation (Figure 8) shows that most of the isolates harboring mycoviruses in F. velutipes do not influence fruiting body development. All the virus-harbouring isolates except for Isolate No.4 produced fruiting bodies. The colony of strain No.4 showed very slow growth, flat morphology and incompatibility as well as fruiting body productivity is lost. These characteristics are similar to the hypovirulence identified in pathogenic fungi [14]. dsRNA of mycovirus in this degenerate wild strain (designated as F. velutipes isometric virus; FvIV), has been sequenced and deposited in DDBJ under the accession no. of AB428575. FvIV is studied in detail in another study and submitted for publication.
The color of the fruiting body, as can be seen in the photograph, shows that virus-free F. velutipes fruiting bodies are whiter (Figure 8). Based on the results of the RT-PCR assay and fruiting body production, the presence of FvBV in mycelium and the dark brown color of fruiting bodies were clearly correlated.
In A. bisporus, brown discoloration of fruiting body caused by bacterial or fungal infection is known [15, 16]. Toxins produced by the bacteria can induce brown coloration of A. bisporus[17]. A quantitative trait locus of A. bisporus resistance to P. tolassi is closely linked to natural cap color [18].
Conclusions
Partitivirus in fungi has been associated with only symptomless infections [11]. For example, a partitivirus in oyster mushroom does not affect its host's phenotype [19]. In this respect, FvBV is possibly unique because it is associated with a specific symptom of F. velutipe s. Although Koch's postulates have not been established yet, an association between a browning of fruiting body and a partitivirus in F. velutipes became evident in this study.
Methods
Strains
Viral particles were purified from mycelium of F. velutipes strain S6B. Strain S6B is a variant of the cultivated F. velutipes strain Shinano No. 6 (designated as S6) that contained dsRNAs and showed brown discoloration of the fruiting bodies [5]. The 14 wild isolates and 2 cultivated strains used in this study are listed in Table 3.
Culturing
The strains were routinely maintained on MA (2% malt extract, 1.8% agarose). Cultures for extraction of dsRNA were grown in 300-ml Erlenmeyer flasks containing 150 ml of GYM (1% glucose, 0.4% yeast extract, and 1% malt extract) at 24°C, stationary for 3 to 4 weeks.
dsRNA extraction
Viral particles of F. velutipes were precipitated with PEG 8000 and NaCl as described previously [3]. Total RNA from the PEG precipitate suspended in TES (10 mM Tris-HCl, 1 mM EDTA, 0.15 M NaCl, pH 7.0) was isolated using the QIAmp Viral RNA Mini Kit (Qiagen, Chatsworth, CA, USA) according to the manufacturer's instructions. dsRNA for agarose gel electrophoresis was prepared from the viral RNA by DNase I (Promega, Madison, WI, USA) and S1 nuclease (Takara Bio, Ohtsu, Japan) digestion. The nuclease-digested product was precipitated with isopropanol after extraction with Sepasol-RNA I Super (Nacalai Tesque, Kyoto, Japan). Total dsRNA was electrophoresed on agarose gel, stained with ethidium bromide, and observed under UV illumination.
Purification of viral particles
The PEG precipitate of viral particles was subjected to 30-40% sucrose density-gradient centrifugation for 2 h at 100,000 × g at 4°C. After the ultracentrifugation, the sample was fractionated into 2-ml volumes, and 100 μl of each fraction was assessed for the presence of dsRNA using the QIAmp Viral RNA Mini Kit (Qiagen). Fractions containing the dsRNA were collected, and viral particles were confirmed by electron microscope observation using a transmission electron microscope (H-7000; Hitachi, Tokyo) after negative staining with 2% aqueous uranyl acetate. dsRNA was extracted from the purified viral particles using the RNeasy Mini Elute Clean-up Kit (Qiagen).
dsRNA sequence
dsRNA extracted from the purified virions was cloned using the Takara small RNA Cloning Kit (Takara Bio) according to the manufacturer's instructions, with a slight modification: RevaTraAce (Toyobo, Osaka, Japan) was used for cDNA synthesis instead of the M-MLV RTase included in the kit, with the aim of generating a longer cDNA. The cDNA was cloned into the pGEM-T easy vector (Promega, Madison, WI, USA) according to the manufacturer's instructions and transformed into E. coli DH5α (Toyobo). Cloned DNA was PCR amplified using Insert Check Ready (Toyobo). The DNA was sequenced using the BigDye Terminators v1.1 Cycle Sequencing Kit (Applied Biosystems, Foster City, CA, USA) with an ABI Prism 3100 Genetic Analyzer (Applied Biosystems).
Sequence analysis
A PSI-BLAST search [9] was performed with the predicted amino acid sequences of the cDNA generated from dsRNA1 and 2. Sequence alignment was performed using the MAFFT program L9INS-I (available at http://align.bmr.kyushu-u.ac.jp/mafft/online/server/index.html) [12]. Neighbour joining with 1,000-boot strapping was performed and phylogenetic tree was constructed with sequences of bootstrap value above 70%. For visually constructing phylogenetic tree, the MAFFT program on the Web server http://align.genome.jp/mafft/ was used. Sequences used for constructing the phylogenetic tree are summarized in Table 1.
RT-PCR
Primers for RT-PCR were designed from the sequence of the putative CP1. Primers CP-F (5'-GCCCGCATCATTACTGTTCT-3') and CP-R (5'-TATCGGTATCAGGGCCGTAG-3') were designed for the amplification of the 495-bp product from nt 479 to 973 of the CP1 DNA using Primer version 3.0 (Whitehead Institute for Biomedical Science). RT-PCR was performed with PEG-precipitated dsRNA prepared as described above with 20 pmol of both primers and RT-PCR quick Master Mix (Toyobo). The dsRNA was denatured at 100°C for 10 min. and immediately cooled on ice. Reverse transcription was performed as follows: 90°C for 30 sec, 60°C for 30 min, and 94°C for 1 min. The following PCR amplification was 40 cycles at 94°C for 30 sec, 55°C for 30 sec, 72°C for 1 min with a final extension of 72°C for 7 min. PCR products were electrophoresed on agarose gels, stained with ethidium bromide, and observed under UV illumination.
Fruiting body production
Fruiting bodies were produced using sawdust supplemented with rice bran as the substrate, contained in polypropylene bottles of 800 ml as described in [4].
References
Hollings M: Viruses associated with die-back disease of cultivated mushrooms. Nature 1962, 196: 962-965. 10.1038/196962a0
Fletcher JT, Gaze R: Abiotic Disorders. In Mushroom Pest and Disease Control. Edited by: Fletcher JT, Gaze R. London: Manson Publishing Ltd; 2008:166-176.
Magae Y, Hayashi N: Double-stranded RNA and virus-like particles in the edible basidiomycete Flammulina velutipes (Enokitake). FEMS Microbiol Lett 1999, 180: 331-335. 10.1111/j.1574-6968.1999.tb08814.x
Magae Y, Nakamura K, Akahane K, Tsunoda S: A simple colorimetric method for detecting degenerate strains of the cultivated basidiomycete Flammulina velutipes (Enokitake). Appl Environ Microbiol 2005, 71: 6388-6389. 10.1128/AEM.71.10.6388-6389.2005
Magae Y: Double-stranded RNA elements in the cultivated Flammulina velutipes , Shinano No.6. Mushroom Sci Biotechnol 2003, 11: 93-96. in Japanese
Grogan HM, Adie BA, Gaze RH, Challen MP, Mills PR: Double-stranded RNA elements associated with the MVX disease of Agaricus bisporus . Mycol Res 2003, 107: 147-154. 10.1017/S0953756203007202
Sonnenberg AS, Lavrijssen : Browning of mushrooms and the presence of viral double-stranded RNA in Dutch mushrooms. In Science and Cultivation of Edible and Medicinal Fungi. Edited by: Romaine CP, Keil CB, Rinker DL, Royse DJ. Penn State, USA; 2004:541-546.
Rao JR, Nelson DW, McClean S: The enigma of double-stranded RNA (dsRNA) associated with mushroom virus X (MVX). Curr Issues Mol Biol 2007, 9: 103-121.
Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ: Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acid Res 1997, 25: 389-402. 10.1093/nar/25.17.3389
Marchler-Bauer A, Anderson JB, Chitsaz F, Derbyshire MK, DeWeese-Scott C, Fong JH, Geer LY, Geer RC, Gonzales NR, Gwadz M, He S, Hurwitz DI, Jackson JD, Ke Z, Lanczycki CJ, Liebert CA, Liu C, Lu F, Lu S, Marchler GH, Mullokandov M, Song JS, Tasneem A, Thanki N, Yamashita RA, Zhang D, Zhang N, Bryant SH: CDD: specific functional annotation with the Conserved Domain Database. Nucleic Acids Res 2009, 37(D): 205-210. 10.1093/nar/gkn845
Ghabrial SA, Ochoa WF, Baker TS, Nibert ML: Partitiviruses. In Encyclopedia of Virology. Edited by: Mahy BWJ, Van Regenmortel MHV. Oxford, Elsevier; 2009:68-75.
Katoh K, Kuma K, Toh H, Miyata M: MAFFT version 5: improvement in accuracy of multiple sequence alignment. Nucleic Acid Res 2005, 33: 511-518. 10.1093/nar/gki198
Bruenn JA: A closely related group of RNA-dependent RNA polymerases from double-stranded RNA viruses. Nucleic Acid Res 21: 5667-5669. 10.1093/nar/21.24.5667
Nuss DL: Hypovirulence: mycoviruses at the fungal-plant interface. Nat Rev Microbiol 2005, 8: 632-642. 10.1038/nrmicro1206
Jolivet S, Arpin N, Wichers HJ, Pellon G: Agaricus bisporus browning: a review. Mycol Res 1998, 102: 1459-1483. 10.1017/S0953756298006248
Soler-Rivas C, Arpin N, Olivier JM, Wichers HJ: Discoloration and tyrosinase activity in Agaricus bisporus fruit bodies infected with various pathogens. Mycol Res 2000, 104: 351-356.
Andolfi A, Cimmino A, Lo Cantore P, Iacobellis NS, Evidente A: Bioactive and Structural Metabolites of Pseudomonas and Burkholderia Species Causal Agents of Cultivated Mushrooms Diseases. Perspect Medicin Chem 2008, 2: 81-112.
Moquet F, Desmerger C, Mamoun M, Ramos-Guedes-Lafargue M, Olivier JM: A quantitative trait locus of Agaricus bisporus resistance to Pseudomonas tolaasii is closely linked to natural cap color. Fungal Genet Biol 1999, 28: 34-42. 10.1006/fgbi.1999.1157
Lim WS, Jeong JH, Jeong RD, Yoo YB, Yie SW, Kim KH: Complete nucleotide sequence and genome organization of a dsRNA partitivirus infecting Pleurotus ostreatus . Virus Res 2005, 108: 111-119. 10.1016/j.virusres.2004.08.017
Shamoun SF, Varga AM, Valverde RA, Ramsfield T, Sumampong G, Elliott M, Masri S, James D: Identification and molecular characterization of a new dsRNA virus infecting Chondrostereum purpureum . Can J Plant Pathol 2008, 30: 604-613. 10.1080/07060660809507561
Osaki H, Nomura K, Iwanami T, Kanematsu S, Okabe I, Matsumoto N, Sasaki A, Ohtsu Y: Detection of a double-stranded RNA virus from a strain of the violet root rot fungus Helicobasidium mompa Tanaka. Virus Gen 2002, 25: 139-145. 10.1023/A:1020105701017
Vainio EJ, Korhonen K, Tuomivirta TT, Hantula J: A novel putative partitivirus of the saprotrophic fungus Heterobasidion ecrustosum infects pathogenic species of the Heterobasidion annosum complex. Fungal Biol 2010, 114: 955-965. 10.1016/j.funbio.2010.09.006
Blawid R, Stephan D, Maiss E: Molecular characterization and detection of Vicia cryptic virus in different Vicia faba cultivars. Arch Virol 2007, 152: 1477-1488. 10.1007/s00705-007-0966-5
Szego A, Enünlü N, Deshmukh SD, Veliceasa D, Hunyadi-Gulyás E, Kühne T, Ilyés P, Potyondi L, Medzihradszky K, Lukács N: The genome of Beet cryptic virus 1 shows high homology to certain cryptoviruses present in phylogenetically distant hosts. Virus Gen 2010, 40: 267-27. 10.1007/s11262-009-0432-4
Boccardo G, Candresse T: Complete sequence of the RNA1 of an isolate of White clover cryptic virus 1, type species of the genus Alphacryptovirus. Arch Virol 2005, 150: 399-402. 10.1007/s00705-004-0422-8
Acknowledgements
This work was supported by the Agriculture, Forestry and Fisheries Research Council of Japan, as part of project for utilizing advanced technologies in agriculture, forestry and fisheries. We are grateful to Mr. Kobun Akahane and Mr. Shigeuyki Tsunoda of Nagano prefecture, for professional cultivation of F. velutipes.
Author information
Authors and Affiliations
Corresponding author
Additional information
Competing interests
The authors declare that they have no competing interests.
Authors' contributions
YM purified the virus, isolated dsRNA, did the phylogenetic analysis and wrote the manuscript. MS cloned the sequences, designed RT-PCR primers and performed the RT-PCR analysis. All authors read and approved the final manuscript.
Authors’ original submitted files for images
Below are the links to the authors’ original submitted files for images.
Rights and permissions
This article is published under license to BioMed Central Ltd. This is an Open Access article distributed under the terms of the Creative Commons Attribution License (http://creativecommons.org/licenses/by/2.0), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
About this article
Cite this article
Magae, Y., Sunagawa, M. Characterization of a mycovirus associated with the brown discoloration of edible mushroom, Flammulina velutipes. Virol J 7, 342 (2010). https://doi.org/10.1186/1743-422X-7-342
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/1743-422X-7-342