Abstract
Background
Rice and maize dwarf diseases caused by the newly introduced Southern rice black-streaked dwarf virus (SRBSDV) have led to severe economic losses in South China in recent years. The distribution and diversity of SRBSDV have not been investigated in the main rice and maize growing areas in China. In this study, the distribution of SRBSDV in China was determined by using reverse transcription-polymerase chain reaction (RT-PCR).
Results
Between 2009 and 2010, 2404 plant samples (2294 rice, 110 maize samples, and more than 300 cultivars) with dwarf symptoms were collected from fields in 194 counties of 17 provinces in China and SRBSDV was detected. The results indicated that 1545 (64.27%) of samples (both rice and maize) were infected with SRBSDV. SRBSDV was detected widely in Hainan, Guangdong, Guangxi, Yunnan, Guizhou, Chongqing, Fujian, Jiangxi, Hunan, Hubei, Anhui, Jiangsu, and Zhejiang provinces, which suggests SRBSDV is an important pathogen causing rice dwarfing diseases in South China. Phylogenetic analysis of 15 representative virus isolates revealed that SRBSDV isolates in China had high levels of nucleotide and amino acid sequence identities (>97.8%).
Conclusions
SRBSDV spreads naturally in Yangtze River basin and south region, the location of the major rice production areas. In comparison, the virus rarely spreads north of Yangtze River in North China. Distribution of SRBSDV is consistent with the migrating and existing ranges of its vector WBPH, suggesting that SRBSDV might be introduced into South China along with the migration of viruliferous WBPH.
Similar content being viewed by others
Background
Southern rice black-streaked dwarf virus (SRBSDV) was first identified in Yangjiang, Guangdong province in China in 2001, and has been proposed as a new member of the genus Fijivirus in the family Reoviridae[1]. Infected rice plants show typical dwarf symptoms, along with dark greening of the leaves, pronounced stunting, twisting of leaf tips, and small white waxy galls along veinlets on the underside of leaf blades and culms [1, 2]. SRBSDV can infect rice causing distinct symptoms at different growth stages, and it can also infect maize (Zea mays), Coix chinensis, Echinochloa, Juncellus and Pennisetum[3]. SRBSDV is transmitted mainly by the white back planthopper (WBPH, Sogatella furcifera Horváth) in a persistent, circulative-propagative manner. The virus, however, cannot be transmitted from WBPH female adults to their progeny via eggs [1]. The small brown planthopper (SBPH, Laodelphax striatellus Fallén), a major vector of the Rice black-streaked dwarf virus (RBSDV) [4], which is most closely related to SRBSDV in phylogeny, can acquire SRBSDV but not transmit it [5]. Available data also shows that WBPH is the major vector of SRBSDV with high efficiency of transmission in rice fields [1]. Moreover, the virus is not transmitted through seed [6].
SRBSDV shares major similarities of symptoms, host ranges, virion morphology, and serology with RBSDV [1, 2], another member of the genus Fijivirus. RBSDV was widespread and prevalent in most parts of East, North, Northwest, and Northeast China in the last century [4, 7]. The two viruses are indistinguishable in agarose or polyacrylamide gel electrophoretic profiles of their genome segments [1]. Due to their overlapping plant host ranges and nearly indistinguishable symptoms in their common plant hosts, it is not easy to distinguish these two virus diseases based on visual symptoms. Recently, the complete nucleotide sequences of SRBSDV and RBSDV genomic RNAs were determined. Ten segments of SRBSDV share 60-80% of nucleotide sequence identities with RBSDV’s counterparts [8–10], facilitating accurate identification of the two viruses by using molecular approaches.
SRBSDV causes a devastating disease that threatens rice production in many provinces in China. In 2009, the rice black-streaked dwarf disease caused by SRBSDV resulted in significant losses in late rice production in South China, especially in Hunan, Hubei, Guangxi, and Jiangxi provinces [11, 12]. In 2010, SRBSDV broke out again in these regions. At the same time, SRBSDV also caused severe losses in northern Vietnam, the overwinter location of WBPH [13, 14]. Accurate identification of the virus and its distribution is the first step in designing effective disease control strategies. To date, however, the available information about the occurrence of SRBSDV in China is limited, given that related investigations were only performed within confined geographic areas. In this paper, we collected 2404 rice and maize samples from distinct geographic areas of China between 2009 and 2010, and the occurrence of SRBSDV was detected by using RT-PCR.
Results
Identification and distribution of SRBSDV in China
A novel rice black-streaked dwarf disease caused by SRBSDV, exhibiting similar symptoms with another rice dwarfing disease caused by RBSDV in the field, was investigated and identified in China. A duplex RT-PCR was performed to distinguish accurately the two viruses. The 569-bp fragments were produced from SRBSDV-infected plant samples, whereas the 1119-bp products were amplified from samples infected by RBSDV. No amplified products were detected from healthy control plants (Figure 1A). Between 2009 and 2010, 2404 rice and maize samples with typical dwarf symptoms were collected and identified from 194 counties in 17 provinces in China. The results showed that SRBSDV was detected in 1537 out of 2294 rice and in 8 out of 110 maize samples, and positive samples were from 166 counties of 14 provinces, excluding Shanghai, Shandong and Hebei (Table 1). The total detection rate of SRBSDV was 64.27%. Precisely, the detection rate was 67.00% in rice samples, and 7.27% in maize plants.
RT-PCR identification of SRBSDV and RBSDV isolates from rice and maize samples in China. A: Simultaneous detection of SRBSDV and RBSDV from infected plant samples. Line M, DNA Markers; line 1, RBSDV positive control; line 2, SRBSDV positive control; line 3–24, rice or maize samples from fields; line 25, negative control. B: RT-PCR analysis of SRBSDV outer CP genes. Line M, DNA Markers; Line 1–14, partial SRBSDV isolates.
On the basis of results from molecular diagnosis, the distribution map of SRBSDV in China was plotted (Figure 2). As is shown in Figure 2, the virus disease occurred throughout most parts of Hainan, Guangdong, Guangxi, Fujian, Jiangxi, Hunan, Hubei, and Anhui provinces, and scattered in southeast parts of Yunnan and Guizhou provinces and southwest parts of Zhejiang province. SRBSDV was only found in a few fields in Sichuan, Chongqing, and Jiangsu provinces. In general, SRBSDV distributed throughout South China, especially in the Yangtze River basin and south of the basin, where hybrid rice is primarily cultivated. In comparison, SRBSDV occurred rarely in northern parts of Yangtze River. However, it was remarkable that five virus isolates were detected from a rice field in north (Xinyi) of Jiangsu province, where japonica rice is widely cultivated, and RBSDV is ubiquitous. Our results reveal that SRBSDV can infect not only hybrid rice, but also conventional rice, and the virus can infect almost all rice varieties in South China. All three main rice types in China, japonica rice, glutinous rice and indica rice, can be infected by SRBSDV. However, the vast majority of cultivars with positive test results were indica rice and hybrid rice in the experiment.
Moreover, detection rate of RBSDV in samples was also counted, and it was found that 11.77% of the rice and 50.00% of the maize samples were infected by RBSDV (Table 1). It was remarkable that plant samples co-infected by SRBSDV and RBSDV were not found. The results indicate that SRBSDV is an important pathogen causing rice dwarf disease in South China, while RBSDV is an important pathogen causing maize rough dwarf disease in maize planting areas of China, such as Shandong and Hebei provinces. Although SRBSDV can also cause maize dwarfing, only a few maize samples were infected by SRBSDV in the experiment.
Genetic diversity of SRBSDV isolates in China
In order to assess the genetic diversity of SRBSDV isolates in China, segment 10 open reading frames (ORFs, encoding outer Coat Protein [CP]) from partial virus isolates were amplified with the specific primers S10-F/S10-R (Figure 1B). Fifteen representative SRBSDV isolates from rice and maize were selected and sequenced, with at least one sample from each province. The sequence analysis of the outer CP genes revealed that there were high levels of nucleotide and amino acid sequence identities (>97.8%) among the 13 SRBSDV isolates from rice (Table 2). Furthermore, two SRBSDV isolates (ZJDY2 and ZJDY4) from maize (Zhejiang province) also exhibited high identities (>98.4%) with other isolates from rice. In general, all outer CP genes from different host and geographical isolates exhibited high levels of sequence identities. When these sequences were compared with the sequences reported by other laboratories, including four isolates from Vietnam (GU017742, GU017741, GU017740 and GU017739) and five isolates from China (EU523360, HM114212, HM114213, HM114214 and EU784840), more than 97.7% nucleotide identity was observed (data is not shown).
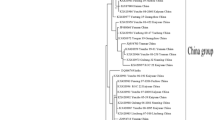
Based on the outer CP gene sequences of SRBSDV isolates detected in the experiment, together with two additional sequences from GenBank (EU784840 and EU523360), the phylogenetic tree was constructed with a RBSDV isolate (AY050489) as the outgroup (Figure 3). No obvious subgrouping could be observed according to hosts or geographical locations of isolates, and all SRBSDV isolates from China were highly similar and clustered in one distinct clade well-separated from the RBSDV isolate. The lower number of nucleotide substitutions per site also showed that there was lower variation among different isolates.
Phylogenetic analysis of SRBSDV based on outer CP gene nucleotide sequence showing the relationships of SRBSDV isolates from different geographic regions in China. Trees were constructed by MEGA 4.0 using the neighbor-joining method with 1000 bootstrap replications (bootstrap values were showed where >50%). Trees were rooted with RBSDV segment 10 (AY050489) as outgroup, and two reported SRBSDV isolates (EU784840, EU523360) were also included as references. Bars indicated numbers of nucleotide substitutions per site.
Discussion
In this paper, the distribution of SRBSDV in China was reported for the first time. Numerous samples from different areas were analyzed for detection of SRBSDV between 2009 and 2010. The results indicate that SRBSDV spreads naturally in the Yangtze River basin and south of the basin, the site of major rice production areas. In comparison, the virus spreads rarely in either north of the Yangtze River or North China, the major sites of maize production, such as Shandong and Hebei provinces.
As a general rule, epidemic and severity of insect-transmission viral diseases on rice depends on the number and viruliferous rate of vector populations, rice varieties, cropping systems, and environment. In 2001, SRBSDV occurred occasionally in Guangdong and Hainan provinces [1]. However, the virus dispersed to neighboring provinces in the next few years and broke out in Hunan and Jiangxi provinces in 2009 [11, 12]. The sudden occurrence and development of SRBSDV on rice in South China between 2009 and 2010 could be attributed to the high viruliferous rate of WBPH populations in the advanced-diseased fields. In advanced-diseased fields, the ratio of viruliferous WBPH was as high as 60% [1].
WBPH is a typical long-distance migration pest, and it is widespread in rice fields in China except in the south of Xinjiang province [15, 16]. Although a low number of WBPH overwinter in Hainan province and in southern regions of Guangxi, Guangdong and Yunnan provinces in China [17], the main source of spring and summer WBPH in China is in the Indo-China Peninsula, primarily in Vietnam, where the Red River Delta is a direct source and the Mekong Delta is an initial source of WBPH [18]. Every year in China, WBPH from Vietnam migrate regularly to the south regions of 29˚ N along with the southwest air flow and land mainly on the south areas of 25˚ N, where they develop and form dominant populations in the local rice fields [15, 17, 18]. Our results show that SRBSDV distributes extensively in the south areas of 29˚ N, which is consistent with the primary landing and existing ranges of WBPH. The previously described WBPH migration pattern suggests that SRBSDV might be introduced into South China along with the migration of the viruliferous WBPH.
Furthermore, SBPH can acquire SRBSDV, but not transmit it [5], which indicates that SBPH cannot play a role in SRBSDV epidemics and dissemination. This is a major cause for rare distribution of SRBSDV in the north of the Yangtze River and North China, where RBSDV transmitted by SBPH has a widespread prevalence. SBPH cannot transmit SRBSDV, while WBPH migrates to the south before winter; therefore, the virus has a very slim chance of existing in the Yangtze River basin and most parts of South China in the winter. Because a few numbers of WBPH can overwinter in Hainan and in the south of Guangxi, Guangdong and Yunnan in China, it was considered that SRBSDV disease might exist in these areas in the winter. This assumption was confirmed after our field trips in the winters of 2011 and 2012. This WBPH winter migration pattern suggests that Hainan and the south of Guangxi, Guangdong and Yunnan are the annual distribution regions of SRBSDV in China and that the rest (Yangtze River basin and most parts of South China) are the seasonal distribution regions of SRBSDV.
The accumulation of viruliferous WBPH in wintering grounds and their migration from their source could explain the fast-spreading of SRBSDV in South China. Our results show that all SRBSDV China isolates have higher identities and lower variation, which indicates that SRBSDV might derive from a single origin. Presently, however, little is known about the origin of SRBSDV and if the virus originates from Vietnam. The SRBSDV disease outbreak in northern Vietnam in 2009 [13, 14] and the virus isolates from China share high levels of nucleotide and amino acid sequence identities with Vietnam isolates [14], but we have no direct evidence to prove whether or not SRBSDV originates in northern Vietnam. This question is important and remains to be further elucidated.
In recent years, an increasing number of inter-subspecies (japonica-indica) hybrid combinations were cultivated and grown to increase rice yield in South China. A previous study indicated that these japonica-indica hybrids were more favorable for WBPH than japonica rice, and the phenomenon was termed as “super-susceptibility” [19]. Other studies suggested that WBPH-resistance genes retained in sympatric japonica rice landraces in China, while there was little ovicidal and sucking-inhibited resistance to WBPH in either hybrid rice or indica rice. The massive introduction and wide cultivation of susceptible hybrid rice provides a favorable environment for WBPH and increases the possibility for the vector to transmit virus, which is a potential threat to rice production in South China. SRBSDV was found in early, middle-season, and late rice, and its infectivity on early rice was less severe than on middle-season and late rice [3]. The migration of 2nd or 3rd generation viruliferous WBPH, together with viral sources accumulated on early rice, might increase the incidence of SRBSDV on middle-season and late rice.
Though limited maize plants were infected by SRBSDV in our investigation, SRBSDV has the potential to cause severe losses in maize. If SRBSDV spreads to North China, where maize is planted widely, through the migration of WBPH, the virus might threaten maize production as RBSDV has done [5, 7]. Notably an isolate of SRBSDV has been obtained from naturally infected maize plants in Shandong province in northern China in 2011 [20], currently the northernmost occurrence of SRBSDV.
In our investigation, all samples had dwarf symptoms, but detection rates of SRBSDV and RBSDV were 64.27% and 13.52% respectively, without co-infection of the two viruses. The remaining samples might be infected by other viruses, such as Rice ragged stunt virus (RRSV), Rice gall dwarf virus (RGDV), Rice dwarf virus (RDV) and Rice grassy stunt virus (RGSV). These viruses are common on rice plants in South China, and can also produce dwarf-like symptoms. Of course, positive samples for SRBSDV might be mixed-infected by these viruses. An expanded surveying for all rice virus categories and distribution is needed in the future, which will be helpful for understanding viral epidemiology and controlling disease.
With increasing global temperature and more frequent warm winters, the attacking period of WBPH might be prolonged, and the vector might advance across Vietnam and across China. In addition to the susceptible varieties and cultivation systems, warmer winters will further aggravate the SRBSDV disease occurrence in rice in South China. Therefore, it is urgent to cultivate resistant rice and maize varieties and develop other integrated control strategies for reducing the possibility of damage by SRBSDV in China.
Materials and methods
Plant samples
Field surveys were done between 2009 and 2010 in 194 counties of 17 provinces in China, including Guangdong, Guangxi, Hunan, Hubei, Jiangxi, Hainan, Yunnan, Guizhou, Sichuan, Chongqing, Fujian, Zhejiang, Shanghai, Jiangsu, Anhui, Shandong, and Hebei provinces. 2404 cereal crop samples (2294 rice and the rest maize samples) with typical stunting or dwarf symptoms were collected from fields in the above mentioned provinces (104˚19′ E to 121˚8′ E, and 19˚25′ N to 34˚38′ N), and these samples covered more than 300 rice and maize cultivars. At least three rice or maize plants were sampled in each field. Leaf or stem tissues of samples were either used directly for total RNA extraction or frozen at −70°C for further study.
RNA extraction and primer design
Total RNAs from healthy and virus-infected control plants as well as field samples were extracted following the standard protocol of TRIzol® reagent (Invitrogen, USA). The concentration and quality of each RNA sample were determined with an Eppendorf Biophotometer plus (Eppendorf, Germany). For the simultaneous detection and distinguishing of SRBSDV and RBSDV, a quick duplex RT-PCR analysis was developed as described by Ji et al. (2011) [21]. Three specific primers (S9-SR-F, S9-RB-F and S9-R) were designed according to the reported segment 9 nucleotide sequences of both viruses (GenBank No. AB011403, AF459812, AY050486, AJ291706, AJ297429, AF536564, AF540976, AY039705, EU523359 and EU784843), which shared 75% identity. Primer S9-R (5’-GGATTACAACAHACACAMCGAAA-3’) was complementary to the nucleotide 1469–1491 of segment 9, which was a conserved region in SRBSDV and RBSDV segment 9. Primer S9-SR-F (5’-TTACAYCAAGCACTTTGCGAGG-3’, corresponding to the nucleotide 923–944 of SRBSDV segment 9), and S9-R were used to amplify a 569-bp fragment, which was specific for SRBSDV. Primer S9-RB-F (5’-GRTAGACAGGCAAAYMTAAGCGT-3’) corresponded to the nucleotide 376–398 of RBSDV segment 9, which could also pair with S9-R to amplify a specific 1119-bp fragment for detection of RBSDV. Moreover, primer S10-F (5’-ATGGCTGACATAAGACTTGACAT-3’), corresponding to the nucleotide 22–44 of SRBSDV segment 10 (GenBank No. EU523360), and S10-R (5’-TCATCTGGTGACTTTATTTAACAC-3’), complementary to the nucleotide 1672–1695 of SRBSDV segment 10 (GenBank No. EU784840), were designed to amplify the virus whole outer Coat Protein (CP) gene (1674-bp), which was used to analyze the genetic diversity among different SRBSDV isolates.
RT-PCR amplification and sequence analysis
First strand cDNA was synthesized with random 6 hexamer as primer by using 1st strand cDNA Synthesis Kit (Invitrogen, USA) according to the manufacturer’s protocols. Subsequent PCR amplification was performed by using Pfu DNA polymerase and designed specific primers. In the duplex RT-PCR, primer S9-SR-F, S9-RB-F and S9-R were mixed into the same reaction system to conduct PCR, and then amplified products were separated using 1.5% agarose gel electrophoresis to distinguish simultaneously SRBSDV and RBSDV. The PCR products from S10-F/S10-R primer were cloned individually using pMD18-T vector system (TaKaRa, Dalian, China) and sequenced using an automated dye terminator sequencing system (model 377; PE Applied Biosystems, USA) according to the manufacturer’s protocol. Sequence data was analyzed with DNAstar software. Phylogenetic analysis and molecular diversity among isolates were estimated by calculating the p-distance values for nucleotide and amino acid comparisons using the MEGA program (version 4.0). To determine the relationships between SRBSDV and RBSDV, a phylogenetic tree was constructed via the neighbor-joining (NJ) algorithm with Kimura 2-parameter model.
References
Zhou GH, Wen JJ, Cai DJ, Li P, Xu DL, Zhang SG: Southern rice black-streaked dwarf virus: a new proposed Fijivirus species in the family Reoviridae. Chin Sci Bull 2008, 53: 3677-3685. 10.1007/s11434-008-0467-2
Zhang HM, Yang J, Chen JP, Adams MJ: A black-streaked dwarf disease on rice in China is caused by a novel Fijivirus. Arch Virol 2008, 153: 1893-1898. 10.1007/s00705-008-0209-4
Zhou GH, Zhang SG, Zou SF, Xu ZW, Zhou ZQ: Occurrence and damage analysis of a new rice dwarf disease caused by Southern rice black-streaked dwarf virus. Plant Prot 2010, 36: 144-146.
Zhou T, Wu LJ, Wang Y, Cheng ZB, Ji YH, Fan YJ, Zhou YJ: Transmission of rice black-streaked dwarf virus from frozen infected leaves to healthy rice plants by small brown planthopper ( Laodelphax striatellus ). Rice Science 2011, 18: 152-156. 10.1016/S1672-6308(11)60022-X
Jia DS, Chen HY, Mao QZ, Liu QF, Wei TY: Restriction of viral dissemination from the midgut determines incompetence of small brown planthopper as a vector of Southern rice black-streaked dwarf virus. Virus Res 2012, 167: 404-408. 10.1016/j.virusres.2012.05.023
Wang K, Zheng JJ, Zhang SG, Zhou GH: Seedling test confirms Southern rice black-streaked dwarf virus cannot be transmitted via rice seed. Guangdong Agric Sci 2010, 37: 95-96.
Wang HD, Chen JP, Wang AG, Jiang XH, Adams MJ: Studies on the epidemiology and yield losses from rice black streaked dwarf disease in a recent epidemic in Zhejiang province, China. Plant Pathol 2009, 58: 815-825. 10.1111/j.1365-3059.2009.02091.x
Zhang HM, Chen JP, Adams MJ: Molecular characterisation of segments 1 to 6 of Rice black-streaked dwarf virus from China provides the complete genome. Arch Virol 2001, 146: 2331-2339. 10.1007/s007050170006
Wang ZH, Fang SG, Xu JL, Sun LY, Li DW, Yu JL: Sequence analysis of the complete genome of Rice black-streaked dwarf virus isolated from maize with rough dwarf disease. Virus Genes 2003, 27: 163-168. 10.1023/A:1025776527286
Wang Q, Yang J, Zhou GH, Zhang HM, Chen JP, Adams MJ: The complete genome sequence of two isolates of Southern rice black-streaked dwarf virus, a new member of the genus Fijivirus. J Phytopathol 2010, 158: 733-737. 10.1111/j.1439-0434.2010.01679.x
Chen Z, Guo R, Zhong L, Qiu G, Chen M, Song B, Liu J, Fan H, Li X, Yang S: Cause of outbreak of Southern rice black-streaked dwarf virus disease at matian township of luxi county, Jiangxi province. Guizhou Agric Sci 2010, 38: 118-120.
Zhang SB, Zhang DY, Liu Y, Luo XW, Cheng FX, Peng ZP, Ma MY: The dwarf disease on rice in Hunan in 2009 was the southern rice black-streaked dwarf disease. Plant Prot 2010, 36: 98-100.
Cuong HV, Hai NV, Man VT, Matsumoto M: Rice dwarf disease in North Vietnam in 2009 is caused by Southern rice black-streaked dwarf virus (SRBSDV). Bull Inst Trop Agr Kyushu Univ 2009, 32: 85-92.
Hoang AT, Zhang HM, Yang J, Chen JP, Hébrard E, Zhou GH, Vinh VN, Cheng JA: Identification, characterization, and distribution of Southern rice black-streaked dwarf virus in Vietnam. Plant Dis 2011, 95: 1063-1069. 10.1094/PDIS-07-10-0535
Hu G: Research progress in migration of rice whitebacked planthopper. Scientia Agricultura Sinica 1981, 14: 25-30.
Otuka A, Watanabe T, Suzuki Y, Matsumura M, Furuno A, Chino M, Kondo T, Kamimuro T: A migration analysis of Sogatella furcifera (Horváth) (Homoptera: Delphacidae) using hourly catches and a three-dimensional simulation model. Agr Forest Entomol 2006, 8: 35-47. 10.1111/j.1461-9555.2006.00284.x
Shen JH, Shang JM, Liu GJ: Management of the whitebacked planthopper, Sogatella furcifera in China: a mini-review. Chin J Rice Sci 2003, 17: 7-22.
Otuka A, Matsumura M, Watanabe T, Dinh VT: A migration analysis for rice planthoppers, Sogatella furcifera (Horváth) and Nilaparvata lugens (Stål) (Homoptera: Delphacidae), emigrating from northern Vietnam from April to May. Appl Entomol Zool 2008, 43: 527-534. 10.1303/aez.2008.527
Zhu ZR, Cheng J, Jiang MX, Zhang XX: Complex influence of rice variety, fertilization timing, and insecticide on population dynamics of Sogatella furcifera (Horváth), Nilaparvata lugens (Stål) (Homoptera: Delphacidae) and their natural enemies in rice in Hangzhou, China. J Pest Sci 2004, 77: 65-74. 10.1007/s10340-003-0024-3
Yin X, Xu FF, Zheng FQ, Li XD, Liu BS, Zhang CQ: Molecular characterization of segments S7 to S10 of a Southern rice black-streaked dwarf virus isolate from maize in northern China. Virol Sin 2011, 26: 47-53. 10.1007/s12250-011-3170-9
Ji YH, Gao RZ, Zhang Y, Cheng ZB, Zhou T, Fan YJ, Zhou YJ: A simplified method for quick detection of Rice black-streaked dwarf virus and Southern rice black-streaked dwarf virus. Chin J Rice Sci 2011, 25: 91-94.
Acknowledgements
This research was supported by the National Key Basic Research and Development Program (973 Program) of China (Grant No. 2010CB126203), the Special Fund for Agro-scientific Research in the Public Interest of China (Grant No. 201003031 and 201303021), the National Natural Science Foundation of China (Grant No. 31000841) and the Natural Science Foundation of Jiangsu Province (Grant No. BK2011270).
Author information
Authors and Affiliations
Corresponding author
Additional information
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
YZ and WL designed the study. ZC, WL, FS, GZ, JW and YZ collected the samples. RG and ZC performed the RT-PCR tests. ZC, SL and YZ analyzed the data. SL, XZ and YZ wrote and finalized the manuscript. All authors read and approved the final manuscript.
Zhaobang Cheng, Shuo Li contributed equally to this work.
Authors’ original submitted files for images
Below are the links to the authors’ original submitted files for images.
Rights and permissions
Open Access This article is published under license to BioMed Central Ltd. This is an Open Access article is distributed under the terms of the Creative Commons Attribution License ( https://creativecommons.org/licenses/by/2.0 ), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
About this article
Cite this article
Cheng, Z., Li, S., Gao, R. et al. Distribution and genetic diversity of Southern rice black-streaked dwarf viru s in China. Virol J 10, 307 (2013). https://doi.org/10.1186/1743-422X-10-307
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/1743-422X-10-307