Abstract
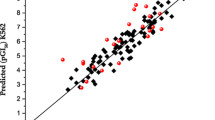
In the present study, we mainly focused on new synthesized 1,7-diazacarbazole derivatives (44 active molecules) as Chk1 inhibitors to build 3D-QSAR model. Comparative molecular field analysis (CoMFA) model with three principal components was developed. The relative contributions in building of CoMFA model were 64.41 % for steric field and 35.59 % for electrostatic field. R 2 values for training and test sets of CoMFA model were 0.8724 and 0.7818, respectively, and squared correlation coefficient for leave-one-out cross-validation test (q 2) was 0.6753. To improve the predictive power, a new 3D-QSAR model was developed by using radial basis function network (RBFN) and score of CoMFA interactions energy values as input variables. Scores 1, 2 and 3 were used as input variables, and a RBFN model with seven centers and spread value equal to 95 was developed to create a nonlinear 3D-QSAR model. R 2 values for training and test sets were 0.9613 and 0.8564, and q 2 for leave-one-out cross-validation test was 0.9258. Docking of all molecules to 3DX ligand binding site of Chk1 receptor indicated six interactions as pharmacological interactions between compounds and binding site of receptors. These pharmacological interactions were hydrogen bonding with LEU-15 and GLU-85 in main chain and four van der Waals interactions with LEU-15, VAL-23, TYR-86 and LEU-137 in side chain. CoMFA contour plots were used to design new inhibitors, and inhibitory activity of each compound was predicted by using CoMFA and RBFN models.









Similar content being viewed by others
References
M. Patil, N. Pabla, Z. Dong, Cell. Mol. Life Sci. 70, 4009 (2013)
H. Hénon, S. Messaoudi, F. Anizon, B. Aboab, N. Kucharczyk, S. Léonce, R.M. Golsteyn, B. Pfeiffer, M. Prudhomme, Eur. J. Pharmacol. 554, 106 (2007)
P.D. Lyne, P.W. Kenny, D.A. Cosgrove, C. Deng, S. Zabludoff, J.J. Wendoloski, S. Ashwell, J. Med. Chem. 47, 1962 (2004)
E. Conchon, F. Anizon, B. Aboab, R.M. Golsteyn, S. Léonce, B. Pfeiffer, M. Prudhomme, Bioorg. Med. Chem. 16, 4419 (2008)
N.-H. Lin, P. Xia, P. Kovar, C. Park, Z. Chen, H. Zhang, S.H. Rosenberg, H.L. Sham, Bioorg. Med. Chem. Lett. 16, 421 (2006)
L. Gazzard, B. Appleton, K. Chapman, H. Chen, K. Clark, J. Drobnick, S. Goodacre, J. Halladay, J. Lyssikatos, S. Schmidt, S. Sideris, C. Wiesmann, K. Williams, P. Wub, I. Yen, S. Malek, Bioorg. Med. Chem. Lett. 24, 5704 (2014)
V. Oza, S. Ashwell, P. Brassil, J. Breed, C. Deng, J. Ezhuthachan, H. Haye, C. Horn, J. Janetka, P. Lyne, N. Newcombe, L. Otterbien, M. Pass, J. Read, S. Roswell, M. Su, D. Toader, D. Yu, Y. Yu, A. Valentine, P. Webborn, A. White, S. Zabludoff, X. Zheng, Bioorg. Med. Chem. Lett. 20, 5133 (2010)
V. Oza, S. Ashwell, P. Brassil, J. Breed, J. Ezhuthachan, C. Deng, M. Grondine, C. Horn, D. Liu, P. Lyne, N. Newcombe, M. Pass, J. Read, M. Su, D. Toader, D. Yu, Y. Yu, S. Zabludoff, Bioorg. Med. Chem. Lett. 22, 2330 (2012)
Y. Tong, M. Przytulinska, Z.-F. Tao, J. Bouska, K.D. Stewart, C. Park, G. Li, A. Claiborne, P. Kovar, Z. Chen, P.J. Merta, M.-H. Bui, A. Olson, D. Osterling, H. Zhang, H.L. Sham, S.H. Rosenberg, T.J. Sowin, N.-H. Lin, Bioorg. Med. Chem. Lett. 17, 5665 (2007)
E. Yuriev, M. Agostino, P.A. Ramsland, J. Mol. Recogn. 24, 149 (2011)
E. Yuriev, P.A. Ramsland, J. Mol. Recogn. 26, 215 (2013)
S.F. Sousa, P.A. Fernandes, M.J. Ramos, Proteins 65, 15 (2006)
G. Sabbagh, N. Berakdar, J. Mol. Graph. Model. 61, 214 (2015)
E. Nazarshodeh, F. Shiri, J.B. Ghasemi, J. Iran. Chem. Soc. 12, 1945 (2015)
S. Pirhadi, F. Shiri, J.B. Ghasemi, J. Iran. Chem. Soc. 11, 1329 (2014)
J.B. Ghasemi, E. Nazarshodeh, H. Abedi, J. Iran. Chem. Soc. 12, 1789 (2015)
V.K. Vyas, M. Ghate, SAR QSAR Environ. Res. 24, 625 (2013)
W. Yang, M. Shu, Y. Wang, R. Wang, Y. Hu, L. Meng, Z. Lin, J. Mol. Struct. 1054–1055, 107 (2013)
Y. Ji, M. Shu, Y. Lin, Y. Wang, R. Wang, Y. Hu, Z. Lin, J. Mol. Struct. 1045, 35 (2013)
A. Balupuri, P.K. Balasubramanian, C.G. Gadhe, S.J. Cho, SAR QSAR Environ. Res. 25, 651 (2014)
S. Hu, H. Yu, L. Zhao, A. Liang, Y. Liu, H. Zhang, Med. Chem. Res. 22, 4992 (2013)
R.W. Wang, L. Zhou, Z. Zuo, X. Ma, M. Yang, Mol. Simul. 36, 87 (2010)
L. Gazzard, K. Williams, H. Chen, L. Axford, E. Blackwood, B. Burton, K. Chapman, P. Crackett, J. Drobnick, C. Ellwood, J. Epler, M. Flagella, E. Gancia, M. Gill, S. Goodacre, J. Halladay, J. Hewitt, H. Hunt, S. Kintz, J. Lyssikatos, C. Macleod, S. Major, G. Médard, R. Narukulla, J. Ramiscal, S. Schmidt, E. Seward, C. Wiesmann, P. Wu, S. Yee, I. Yen, S. Malek, J. Med. Chem. 58, 5053 (2015)
HyperChem Release 7.1 for windows molecular modeling system program package, HyperCube, 2002
P. Tosco, T. Balle, J. Mol. Model. 17, 201 (2011)
J.B. Ghasemi, F. Shiri, Med. Chem. Res. 21, 2788 (2012)
J.-M. Yang, C.-C. Chen, Proteins 55, 288 (2004)
J.-M. Yang, J. Comput. Chem. 25, 843 (2004)
K.-C. Hsu, Y.-F. Chen, S.-R. Lin, J.-M. Yang, BMC Bioinformatics 12(Suppl 1), S33 (2011)
P. Tosco, T. Balle, F. Shiri, J. Comput. Aided Mol. Des. 25, 777 (2011)
N.J. Richmond, P. Willett, R.D. Clark, J. Mol. Graph. Model. 23, 199 (2004)
G. Cruciani, Molecular Interaction Fields: Applications in Drug Discovery and ADME Prediction (Wiley, Weinheim, 2006)
M.A. Kastenholz, M. Pastor, G. Cruciani, E.E.J. Haaksma, T. Fox, J. Med. Chem. 43, 3033 (2000)
M. Pastor, G. Cruciani, S. Clementi, J. Med. Chem. 40, 1455 (1997)
M. Baroni, G. Costantino, G. Cruciani, D. Riganelli, R. Valigi, S. Clementi, Quant. Struct.-Act. Relat. 12, 9 (1993)
M. Baroni, S. Clementi, G. Cruciani, G. Costantino, D. Riganelli, J. Chemom. 6, 347 (1992)
M. Andersson, J. Chemom. 23, 518 (2009)
S. Wold, M. Sjöström, L. Eriksson, Chemom. Intell. Lab 58, 109 (2001)
G. Cruciani, M. Baroni, S. Clementi, G. Costantino, D. Riganelli, B. Skagerberg, J. Chemom. 6, 335 (1992)
Z. Hassanzadeh, M. Kompany-Zareh, R. Ghavami, S. Gholami, A. Malek-Khatabi, J. Mol. Struct. 1098, 191 (2015)
A. Malek-Khatabi, M. Kompany-Zare, S. Gholami, S. Bagheri, Chemom. Intell. Lab 135, 157 (2014)
J.P. Doucet, A. Panaye, Three Dimensional QSAR: Applications in Pharmacology and Toxicology (Taylor and Francis, London, 2010)
A. Golbraikh, A. Tropsha, J. Mol. Graph. Model. 20, 269 (2002)
G. Melagraki, A. Afantitis, Chemom. Intell. Lab 123, 9 (2013)
G. Melagraki, A. Afantitis, RSC Adv. 4, 50713 (2014)
P.K. Ojha, I. Mitra, R.N. Das, K. Roy, Chemom. Intell. Lab 107, 194 (2011)
A. Tropsha, Mol. Inf. 29, 476 (2010)
D. Wang, Y. Yuan, S. Duan, R. Liu, S. Gu, S. Zhao, L. Liu, J. Xu, Chemom. Intell. Lab 143, 7 (2015)
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Sepehri, B., Hassanzadeh, Z. & Ghavami, R. Pharmacophore interactions analysis and prediction of inhibitory activity of 1,7-diazacarbazoles as checkpoint kinase 1 inhibitors: application of molecular docking, 3D-QSAR and RBF neural network. J IRAN CHEM SOC 13, 1525–1537 (2016). https://doi.org/10.1007/s13738-016-0869-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13738-016-0869-z