Abstract
The cheese microbial ecosystem is complex, and the presence of non-starter adventitious microorganisms in milk may have an influence on the organoleptic characteristics of cheese. The aim of this study was to analyze the composition and diversity of the fungal flora of raw milk destined for cheesemaking from 19 dairy farms in Quebec and to monitor their evolution throughout ripening. Six hundred ten yeast and mold isolates were collected from raw milk and raw milk cheeses over a 9-month period. Based on the sequences of the rDNA ITS1-5.8S-ITS2 region, 67% of the raw milk isolates were yeasts, which were assigned to 37 species across 11 genera, while 33% were molds, which were assigned to 33 species across 25 genera. A semi-quantitative analysis of the yeasts and molds in the raw milk from four farms was performed over a 5-month period. The composition and diversity of the fungal microflora were totally different for each farm, each of which had a unique species profile. To determine whether adventitious yeast strains from the milk could develop in raw milk cheese, a multilocus-sequence-typing (MLST) analysis was performed on 13 Issatchenkia orientalis (syn. Pichia kudriavzevii, anamorph: Candida krusei) isolates. The same MLST genotypes were identified for strains independently isolated from raw milk and raw milk cheese from a farm processing its own milk. This study contributes to the understanding of the natural fungal microflora of raw milk and suggests that non-starter yeasts and molds can transfer from raw milk to raw milk cheese and may influence cheese ripening.
魁北克生乳及特色干酪中真菌群落的特性
摘要 干酪微生物生态系统的复杂性影响了干酪的感官特性。原料乳中外来微生物作为非发酵剂能够影响干酪的成熟。对来自魁北克牧场19个生乳样品中真菌的组成和多样性及其加工的干酪中真菌生长繁殖情况进行了分析和评价。在9个月的实验期间,从生乳及干酪中共分离出了610株酵母和霉菌。根据rDNA ITS1-5.8S-ITS2氨基酸序列分析,酵母占总菌株的67%,共有11个属,37个种。霉菌占33%,共有25个属,33个种。在五个月的实验期间,对四个牧场生乳中的酵母和霉菌进行了半定量分析,结果表明每个牧场样品间真菌群落的组成和多样性差异显著,每个牧场都有其独特的真菌菌群特性。为了分析原料生乳中外来酵母菌株是否能在干酪中生长繁殖,对13株东方伊萨酵母(库德里阿兹威氏毕赤酵母)(Pichia kudriavzevii)进行了多位点序列分型(MLST)分析。并对从来自一个牧场的生乳及其干酪中单独分离出的菌株进行了相似多位点序列分型基因的鉴定。该研究有助于了解生乳中天然真菌群,同时也说明了非发酵剂酵母和霉菌能够从原料生乳中转移到干酪中并影响干酪的成熟。
Similar content being viewed by others
Avoid common mistakes on your manuscript.
1 Introduction
Cheeses harbor a complex microbial ecosystem that evolves during ripening. The coexistence of microorganisms with different activities determines the organoleptic characteristics of cheeses (Arfi et al. 2005). This ecosystem is essential since aseptic cheeses lack the aromatic compounds produced during ripening (Perry and McGillivray 1964). Controlling the ripening process requires in-depth knowledge of the succession of microbial communities and their activities in cheese. In addition to the starter culture, uninoculated (adventitious) microorganisms can participate in the cheese ripening (Swearingen et al. 2001). Non-starter lactic acid bacteria (NSLAB) in cheddar cheese are a good example of this (Beresford and Williams 2004). The characterization of NSLAB, mainly lactobacilli, has revealed that they make a positive contribution to the ripening of a number of cheeses (Law 2010; Swearingen et al. 2001), which has led to the development of commercial adjunct cultures to accelerate cheese ripening (Settanni and Moschetti 2010).
Studies have also shown that adventitious yeasts and molds can contaminate milk and cheese and contribute to the ripening of specialty cheeses (Mounier et al. 2006; Petersen et al. 2002; Vacheyrou et al. 2011). Potential sources of contamination include ambient air, farm workers, cheesemakers, brine, and cheesemaking equipment (Borelli et al. 2006; Mounier et al. 2006; Pereira-Dias et al. 2000; Seiler and Busse 1990; Simov et al. 2006; Vacheyrou et al. 2011; Viljoen et al. 2003). Over 40 non-starter yeasts and molds (NSYM) species have been isolated from milk, brines and cheeses, the most common being Candida catenulata, Candida intermedia, Candida lusitaniae, Candida parapsilosis, Candida rugosa, Candida zeylanoides, Cryptococcus curvatus, Issatchenkia orientalis (syn. Pichia kudriavzevii, anamorph: Candida krusei), Kluyveromyces marxianus, Pichia fermentans, Pichia guillermondii, Saccharomyces cerevisiae, Torulaspora delbrueckii (anamorph: Candida colliculosa), and Yarrowia lipolytica (Baroiller and Schmidt 1990; Borelli et al. 2006; Callon et al. 2007; El-Sharoud et al. 2009; Fadda et al. 2004; Jacques and Casaregola 2008; Lopandic et al. 2006; Roostita and Fleet 1995; Rohm et al. 1992; Seiler and Busse 1990; Seker 2010; Sohier et al. 2009). The main non-starter mold species in dairy products are Cladosporium cladosporioides, Epicoccum nigrum, Mucor racemosus, Penicillium brevicompactum, Penicillium chrysogenum, and Penicillium commune (Hocking and Faedo 1992; Montagna et al. 2004; Samson et al. 2004). While some of these species are known to be undesirable (e.g., Cladosporium cladosporioides, M. racemosus, Penicillium commune), the impact of most NSYM in cheeses is unknown. It has, however, been suggested that they may make important contributions to the flavor and texture of specialty cheeses (Fleet 1990; Pereira-Dias et al. 2000).
While milk may be a source of NSYM, little is known about the composition and diversity of the fungal microflora of milk. A study of Chinese milk reported the presence of 11 yeast species but did not address mold diversity (Chen et al. 2010). Little is known about fungal diversity in raw cow milk. No molecular analyses of fungal species in cheese or studies on changes in yeast and mold diversity over time of milk from dairy farms that produce their own cheeses have been published to our knowledge.
Over 300 cheese varieties are produced in Canada, 53% of which come from the province of Quebec (Statistics Canada 2010). Most varieties are produced by artisan cheesemakers, some of whom process the milk from their own dairy cows to better control the quality of the milk and cheese. The aim of the present study was to analyze the fungal composition and diversity of milk from 19 dairy farms in the province of Quebec. Yeasts and molds in 111 raw milk samples were isolated and characterized. A more detailed analysis of the raw milk and cheese from four dairy farms over a 5-month period was also conducted to characterize their specific microflora and how it varied over time. A multilocus sequence typing (MLST) analysis of adventitious yeast I. orientalis isolates (anamorph: Candida krusei) was also performed to determine whether NSYM from raw milk could be transferred to raw cheese.
2 Materials and methods
2.1 Selection of dairy farms and sampling of cow milk and cheeses
Nineteen dairy farms in six different administrative regions of the province of Quebec were selected. The farms produced specialty cheeses from the cow milk they produced. The names and locations of the farms are being withheld for confidentiality purposes. One hundred eleven raw milk samples were collected between January and November 2009. The major yeast and mold species in each sample were isolated and identified. The samples were collected monthly in each farm directly from bulk tanks using sterile 1-L bottles. Depending from the size of the farm, milk obtained could be the result of one or more milking. The bottles were placed on ice and were sent to the laboratory as quickly as possible. While most samples were analyzed within 24 h, some took 48 h due to travel time constraints. Others were discarded because they took longer than 48 h to arrive or the temperature of the sample was too high on arrival. The farms also provided specialty cheeses for analysis (Table 1). The cheeses were allowed to ripen for 7–10 days before the samples were sent to the laboratory where NSYM flora was analyzed. Samples were aseptically removed from the rind (9 cm2 on each face) and core (11 g).
2.2 Isolation of yeasts and molds from milk and specialty cheese samples
The milk samples were serially diluted in sterile 0.1% (w/v) peptone solution and were plated on YEGC agar medium (10 g.L−1 yeast extract, 10 g.L−1 glucose, 0.1 g.L−1 chloramphenicol, 15 g.L−1 agar) (Guichard and Bonnarme 2005). The rind and core samples (11 g) were placed in 99 mL of sterile 2% (w/v) sodium citrate preheated to 45 °C and were homogenized using a Stomacher lab blender (Seward Stomacher® 400 Circulator) for 90 s at 250 rpm (Lopandic et al. 2006). Serial dilutions in peptone solution were prepared and spread on YEGC agar plates. The plates were incubated at 25 °C for 3 days. Individual colonies were selected based on shape and color, assuming that colonies with the same morphology belonged to the same genus or species. The colonies were re-streaked on YEGC agar plates, which were incubated at 25 °C for 3 days and then kept at 4 °C until the colonies were identified. Samples of the isolates were also suspended in YEGC broth containing 10% (v/v) glycerol and stored at −80 °C. A total of 608 strains, 505 from the milk samples and 103 from the cheese samples, were isolated.
2.3 Semi-quantitative analysis of fungal microflora of milk from four dairy farms
Four dairy farms (A, B, C, and D) were selected for a semiquantitative analysis of the fungal microflora of the milk. Milk samples from each farm were collected once a month from June to October 2009. The samples were diluted, plated, and incubated as described above. Since selecting colonies based on morphology alone can introduce bias, every colony was systematically purified and identified. Five to 25 colonies were isolated, depending on the sample, for a total of 272 isolates. The purified isolates were grown on YEGC agar plates and stored at 4 °C until the isolates were characterized. For long-term storage, samples were suspended in YEGC broth containing 10% (v/v) glycerol and were stored at −80 °C. The semiquantitative assessment was based on the proportion of each species identified and the total yeast and mold counts in each sample.
2.4 DNA isolation and sequencing of the ITS1-5.8S-ITS2 rDNA region
Genomic DNA was isolated according to the method of Al-Samarrai and Schmid (2000). A ∼550-pb fragment from each isolate was PCR amplified using the universal ITS1 (5′- TCC GTA GGT GAA CCT GCG G - 3′) and ITS4 (5′- TCC TCC GCT TAT TGA TAT GC -3′) primers flanking the ITS1-5.8S-ITS2 ribosomal DNA (rDNA) region. The 50-μL PCR reaction mixtures contained 2 U of Taq DNA polymerase (New England Biolab), 1× Taq buffer (10 mM Tris–HCl, 50 mM KCl, 1.5 mM MgCl2, pH 8.3), 100 μM dNTP (GE Healthcare), 50 μM of each primer (ITS1 and ITS4), and 50 ng of DNA template. The PCR assays were performed using a PTC-200 DNA Engine Thermal Cycler (BioRad). Samples were incubated for 2 min at 94 °C followed by 35 cycles using the following conditions: 30 s at 94 °C, 2 min at 55 °C, 2 min at 72 °C, and a final 5-min extension step at 72 °C. The PCR products were sequenced using the ITS1 and ITS4 primers and a 3130xl genetic analyzer (Applied Biosystems). Sequences were assembled using the Staden software package and compared to public databases using the nucleotide–nucleotide basic local alignment search tool (BLAST).
2.5 Multilocus sequence typing of I. orientalis isolates
Thirteen I. orientalis isolates (anamorph Candida krusei) were analyzed by multilocus sequence typing (MLST) as described previously by Jacobsen et al. (2007). Briefly, six loci (ADE2, HIS3, LEU2, LYS2, NMT1, and TRP1) were sequenced and were compared to the Candida krusei scheme at http://pubmlst.org/ckrusei/. The locus and sequence type (ST) numbering system of the scheme was used when possible. New STs were identified and deposited in the pubmlst.org database.
3 Results and discussion
3.1 Yeast and mold diversity in milk
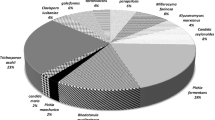
Colonies (505) from 111 cow milk samples were isolated and selected based on differences in shape and color. This approach was selected to increase the probability of isolating a large number of species. The molecular identification approach using the ITS1-5.8S-ITS2 rDNA region revealed that 339 (67%) of the isolates were yeasts that could be assigned to 37 species belonging to 11 genera (Table 2) and that the remaining isolates were molds that could be assigned to 33 species belonging to 25 genera (Table 3).
Of the 339 yeast isolates, Debaryomyces hansenii was the most abundant species (21%) and was detected in the milk from 14 of the 19 dairy farms. Candida was the most abundant genus (44%) and the most diverse, with 148 isolates and 11 species. Cryptoccocus spp. and Pichia spp. made up 10.9% and 8.0% of the yeast isolates, respectively. Of the 37 yeast species identified, 14 were isolated only once. Eurotium was the most abundant mold genus (39 isolates or 23.5%) and was detected in the milk from 13 of the 19 farms. Lichtheimia (Lichtheimia corymbifera) was the second most abundant mold genus, with 22 isolates (13.3%) from nine farms. Fifteen mold species were isolated only once.
Candida glaebosa was the second most frequent yeast isolate and was found in the raw milk from two farms (Table 2). However, 47 of the Candida glaebosa 48 isolates were found in the milk of a single dairy farm (dairy farm A). This species has been identified in cheeses affected by thread mould spoilage but has not been clearly associated with this phenomenon (Hocking and Faedo 1992). These findings indicate that the fungal profile of milk differs from one farm to another. While other investigators have reported that most of the species identified in our study are commonly found in dairy products (Borelli et al. 2006; El-Sharoud et al. 2009; Fadda et al. 2004; Lopandic et al. 2006; Montagna et al. 2004; Pereira-Dias et al. 2000; Prillinger et al. 1999; Roostita and Fleet 1995; Vasdinyei and Deák 2003), only a few have been shown to be part of the native fungal ecosystem of milk.
We also isolated Prototheca zopfii, a yeast-like algal species, from two milk samples. While this non-photosynthetic algal species is thought to be a putative causal agent of bovine mastitis, it has not been possible to make a definite correlation between the presence of Prototheca zopfii and the sub-clinical and clinical symptoms of the disease (Kishimoto et al. 2009; Marques et al. 2010).
Despite the large number of isolates, it is likely that only a fraction of the fungal species in the samples were actually identified. Some species may have had difficulties growing on the synthetic medium or were present in such small numbers that they were masked by the dominant species. However, this overview of the fungal microflora of the milk samples provides an inkling of the complexity of this ecosystem.
3.2 Semiquantitative analysis of fungi in milk from four dairy farms over a 5-month period
To determine whether the fungal ecosystem differed from farm to farm or changed over time for a given farm, the yeast and mold populations in milk samples from four dairy farms were analyzed over a 5-month period. However, rather than selecting colonies based on morphology, every colony was purified and identified by sequencing the ITS1-5.8S-ITS2 rDNA region. This approach was used to eliminate any bias that may have been introduced by selecting colonies based on morphology.
Dairy farm A produced milk with the highest fungal count, with an average of 1.8 × 103 CFU.mL−1 (Online resource 1), while farm c produced milk with the lowest fungal count, with an average of 3.6 CFU.mL−1 (Online resource 2). Yeasts dominated the microflora of the milk from farms A–C, accounting for 91%, 86%, and 82% of the isolates, respectively. Molds dominated the microflora of the milk from farm D, accounting for 67% of the isolates (Online resource 3). While this systematic approach gave a more precise, quantitative portrait of the fungal ecosystem, less species diversity was observed. It also revealed that the dominant species and the composition of the ecosystem differed from one milk sample to another. The samples analyzed over the 5-month period revealed that the milk from farm A was dominated by the fungal species Candida glaebosa, which made up 59% of the isolates. This species was also present in every sample obtained from this farm. Candida zeylanoides and Candida parapsilosis were the dominant species in the milk of farm b, accounting for 31% and 25% of the isolates, respectively (Online resource 4). However, they were only found in three of the five milk samples.
Interestingly, D. hansenii was the dominant yeast species in the milk from farm C. On average, it made up 49% of the total fungal population and was found in four of the five samples. This species is commonly inoculated in dairy products. Some strains have a positive effect on cheese ripening through their proteolytic and lipolytic activities and are able to protect against contaminants (Breuer and Harms 2006; Corsetti et al. 2001). Farm D was unique in that, while D. hansenii (16%) was the predominant yeast species, the microflora of the milk was dominated by a diverse population of mold species (67% of isolates), including Lichtheimia corymbifera (found in all samples), Eurotium spp., Acremonium spp., Aspergillus spp., Cladosporium spp., and Penicillium spp. (Online resource 3).
The composition of the NSYM from milk differs from farm to farm but may also differ along the months. This study also corroborates other studies reporting that the period had a significant effect on the global yeast profile of milk. This could be due to the combined effects of the feeding regime, physiological state of the animal, the weather, and the indoor and outdoor environments (Baroiller and Schmidt 1990; Callon et al. 2007; Zheng et al. 2006). Moreover, cheesemaking process and equipment could be sources of NSYM, especially brine and ripening chambers (Seiler and Busse 1990; Prillinger et al. 1999; Zheng et al. 2006).
3.3 Non-starter yeasts and molds isolated from cheese samples
Non-starter yeasts and molds (103 isolates) were isolated from 28 surface and core samples from 1-week-old specialty cheeses produced from raw and pasteurized milk (Table 1, Table 4). The NSYM colonies were selected based on morphology in order to isolate as many NSYM species as possible. Given that some species are used as ripening cultures, including Penicillium camemberti, Geotrichum candidum (teleomorph Galactomyces candidus), Kluyveromyces lactis, and D. hansenii, isolates corresponding to these species were not included in the analysis since it was impossible to determine whether they were natural contaminants or members of the ripening culture.
Thirty different NSYM species were isolated from the rind and core samples. Of these, 12 species were isolated only once. Yeasts were isolated more frequently than molds, and accounted for 73% of the isolates. Many NSYM isolated from the cheese samples were not found in the milk, including Aspergillus fumigatus, Cochliobolus sativus, Fusarium domesticum, Mucor circinelloides, Cryptococcus diffluens, Pichia membranifaciens, Saccharomyces servazii, Trichosporon asahii, Trichosporon domesticus, and Trichosporon jirovecii. The proportion of these species in the milk samples may have been too low for them to be detected, or they may have been contaminants from the environment, workers, the ripening chamber, or other sources.
The most frequent yeast species isolated was I. orientalis (syn. P. kudriavzevii), which accounted for 25% of the yeast isolates. However, this frequency may have been due to its colonial morphology, which is different from the common white smooth regular colonies often observed with other species such as Candida zeylanoides, D. hansenii, and Candida glaebosa. I. orientalis was reported as a common species found in Graukäse cheese and was also isolated from brines (Prillinger et al. 1999; Seiler and Busse 1990). However, the contribution of this species to cheese ripening process needs to be investigated. M. racemosus was the most frequently isolated mold species. This species is frequently isolated as NSYM and is undesired in most specialty cheeses, such as Camembert (Bekada et al. 2008).
Most of the isolates (59%) came from the core samples. Seven of the 19 yeast species and seven of the 11 mold species from the core samples were not detected in the rind samples (Table 4). Because most cheeses analyzed in this study are surface ripened, the use of ripening culture may have reduced the number of NSYM isolated at the surface. F. domesticum, C. curvatus, Pichia membranifaciens, and Trichosporon aquatile were only isolated from the rind samples. Fewer species were isolated from the rind samples than from the core samples. However, the microflora of the ripening culture may have masked the NSYM species to some extent.
3.4 MLST genotyping of I. orientalis isolates
MLST genotyping was performed on 13 I. orientalis isolates (Table 5) to assess the distribution and persistence of non-starter yeasts in cheese. Leaving aside LMA-694, LMA-696, and LMA-726, two main groups were identified. The first group consisted of LMA-250, LMA-519, LMA-545, LMA-652, and LMA-805, which belong to the St-67 sequence type. The second group was more heterogeneous and was comprised of LMA-422, LMA-783, LMA-546, LMA-503, and LMA-666. The strains in this group shared many loci and all harbored the same ADE1 locus.
Eight MLST profiles were identified in the nine isolates from the milk samples, indicating that at least eight different strains were present. Four of these isolates came from the milk from farm A. LMA-545 and LMA-546 came from the same milk sample but had different MLST profiles.
The allelic St-67 profile encompassed all the isolates from cheese 1 (farm A), despite the fact that the cheese samples were collected from three different lots (April 3, June 12, and August 12), suggesting that this strain or close relatives persisted in the processing plant. Interestingly, the same MLST genotype was found in the raw milk samples from this farm, suggesting that I. orientalis in milk may have survive the cheesemaking process and grow in raw milk cheese. An isolate from cheese sample 2 from dairy farm A had a totally different genotype profile (LMA-503), confirming that several I. orientalis strains can coexist in the same cheesemaking environment and develop in cheeses.
It is noteworthy that LMA-250 and LMA-545 had the same MLST genotype but were isolated from different farms. This suggested that closely related strains could be found in widely separated locations. While the MLST analysis could not guarantee that these two isolates were identical strains, it did suggest a strong genetic affiliation.
4 Conclusion
The NYSM population in milk is very diverse, and the importance of this microflora is often underestimated. The present study provided a clearer picture of the fungal microbiota of milk and confirmed that the diversity and composition of the microbiota can vary from farm to farm. In most cases, yeasts predominated, but some ecosystems were dominated by molds, indicating that the relative proportions of yeasts and molds can vary considerably. The present study also revealed that the fungal ecosystem of milk can remain stable over time. In addition, the MLST genotyping of non-starter yeast isolates strongly suggested that I. orientalis in milk could survive the cheesemaking process and develop in raw milk cheese. Our results also suggested that NSYM can transfer from milk to cheese and influence the raw cheese ripening. Additional studies will be required to determine whether NSYM play a positive or negative role in the development of the cheese flavor.
References
Al-Samarrai TH, Schmid J (2000) A simple method for extraction of fungal genomic DNA. Lett Appl Microbiol 30:53–56
Arfi K, Leclerq-Perlat MN, Spinnler HE, Bonnarme P (2005) Importance of curd-neutralising yeasts on the aromatic potential of Brevibacterium linens during cheese ripening. Int Dairy J 15:883–891
Baroiller C, Schmidt JL (1990) Contribution à l’étude de l’origine des levures du fromage de Camembert. Lait 70:67–84
Bekada AMA, Benakriche B, Hamadi K, Bensoltane A (2008) Modelling of effects of water activity, pH and temperature on the growth rate of Mucor racemosus isolated from soft Camembert cheese. World J Agr Sci 4:790–794
Beresford T, Williams A (2004) The microbiology of cheese ripening. In: Fox PF (ed) Cheese: chemistry, physics and microbiology. Vol. 1, 3rd edn. Elsevier Academic Press, Amsterdam
Borelli BM, Ferreira EG, Lacerda ICA, Franco GR, Rosa CA (2006) Yeast populations associated with the artisanal cheese produced in the region of Serra da Canastra, Brazil. World J Microbiol Biotechnol 22:1115–1119
Breuer U, Harms H (2006) Debaryomyces hansenii—an extremophilic yeast with biotechnological potential. Yeast 23:415–437
Callon C, Duthoit F, Delbes C, Ferrand M, Le Frileux Y, De Crémoux R, Montel M-C (2007) Stability of microbial communities in goat milk during a lactation year: molecular approaches. Syst Appl Microbiol 30:547–560
Chen L-S, Ma Y, Maubois J-L, Chen L-J, Liu Q-H, Guo J-P (2010) Identification of yeasts from raw milk and selection for some specific antioxidant properties. Int J Dairy Tech 63:47–54
Corsetti A, Rossi J, Gobbetti M (2001) Interactions between yeasts and bacteria in the smear surface-ripened cheeses. Int J Food Microbiol 69:1–10
El-Sharoud WM, Belloch C, Peris D, Querol A (2009) Molecular identification of yeasts associated with traditional Egyptian dairy products. J Food Sci 74:341–346
Fadda ME, Mossa V, Pisano MB, Deplano M, Cosentino S (2004) Occurrence and characterization of yeasts isolated from artisanal Fiore Sardo cheese. Int J Food Microbiol 95:51–59
Fleet GH (1990) Yeasts in dairy products. J Appl Bacteriol 68:199–211
Guichard H, Bonnarme P (2005) Development and validation of a plate technique for screening of microorganisms that produce volatile sulfur compounds. Anal Biochem 338:299–305
Hocking AD, Faedo M (1992) Fungi causing thread mould spoilage of vacuum packaged cheddar cheese during maturation. Int J Food Microbiol 16:123–130
Jacobsen MD, Gow AR, Maiden MCJ, Shaw DJ, Odds FC (2007) Strain typing and determination of population structure of Candida krusei by multilocus sequence typing. J Clin Microbiol 45:317–323
Jacques N, Casaregola S (2008) Safety assessment of dairy microorganisms: the hemiascomycetous yeasts. Int J Food Microbiol 126:321–326
Kishimoto Y, Kano R, Maruyama H, Onozaki M, Makimura K, Ito T, Matsubara K, Hasegawa A, Kamata H (2009) 26S rDNA-based phylogenetic investigation of Japanese cattle-associated Prototheca zopfii isolates. J Vet Med Sci 72:123–126
Law B (2010) Cheese adjunct cultures. Aust J Dairy Technol 65:45–49
Lopandic K, Zelger S, Banszky LK, Eliskases FL, Prillinger H (2006) Identification of yeasts associated with milk products using traditional and molecular techniques. Food Microbiol 23:341–350
Marques S, Silva E, Thompson G (2010) Phenotypic characterisation of mastitic Prototheca spp. isolates. Res Vet Sci 89:5–9
Montagna MT, Santacroce MP, Spilotros G, Napoli C, Papa A, Dragoni I (2004) Investigation of fungal contamination in sheep and goat cheese in Southern Italy. Mycopathologia 158:245–249
Mounier J, Goerges S, Gelsomino R, Vancanneyt M, Vandemeulebroecke K, Hoste B, Brennan NM, Scherer S, Swings J, Fitzgerald GF, Cogan TM (2006) Sources of the adventitious microflora of a smear-ripened cheese. J Appl Microbiol 101:668–681
Pereira-Dias S, Potes ME, Marinho A, Malfeito-Ferreira M, Loureiro V (2000) Characterisation of yeast flora isolated from an artisanal Portuguese ewes’ cheese. Int J Food Microbiol 60:55–63
Perry KD, McGillivray WA (1964) The manufacture of ‘normal’ and ‘starter-free’ Cheddar cheese under controlled bacteriological conditions. J Dairy Res 31:155–165
Petersen KM, Westall S, Jespersen L (2002) Microbial succession of Debaryomyces hansenii strains during the production of Danish surfaced-ripened cheeses. J Dairy Sci 85:478–486
Prillinger H, Molnar O, Eliskases-Lechner F, Lopandic K (1999) Phenotypic and genotypic identification of yeasts from cheese. Antonie Van Leeuwenhoek 75:267–283
Rohm H, Eliskases-Lechner F, Bräuer M (1992) Diversity of yeasts in selected dairy products. J Appl Microbiol 72:370–376
Roostita R, Fleet GH (1995) The occurrence and growth of yeasts in Camembert and Blue-veined chesses. Int J Food Microbiol 28:393–404
Samson RA, Hoekstra ES, Frisvad JC (2004) Introduction to food- and airborne fungi, 7th edn. Centralbureau voor Scchimmelcultures, Utrecht
Seiler H, Busse M (1990) The yeast of cheese brines. Int J Food Microbiol 11:289–304
Seker E (2010) Identification of Candida species isolated from bovine mastitic milk and their in vitro haemolytic activity in western turkey. Mycopathologia 169:303–308
Settanni L, Moschetti G (2010) Non-starter lactic acid bacteria used to improve cheese quality and provide health benefits. Food Microbiol 27:691–697
Simov ZI, Simova ED, Beshkola DM (2006) Impact of two starter cultures on proteolysis on Kashkaval cheese. World J Microbiol Biotechnol 22:147–156
Sohier D, Le Dizes A-S, Thuault D, Neuveglise C, Coton E, Casaregola S (2009) Important genetic diversity revealed by inter-LTR PCR fingerprinting of Kluyveromyces marxianus and Debaryomyces hansenii strains from French traditional cheeses. Dairy Sci Technol 89:569–581
Statistics Canada, Agriculture Division, Livestock and Animal Products Section (2010) Dairy statistics, fourth quarter 2009. Statistics Canada, Ottawa, p 44
Swearingen PA, O’Sullivan DJ, Warthesen JJ (2001) Isolation, characterization, and influence of native, nonstarter lactic acid bacteria on cheddar cheese quality. J Dairy Sci 84:50–59
Vacheyrou M, Normand A-C, Guyot P, Cassagne C, Piarroux R, Bouton Y (2011) Cultivable microbial communities in raw cow milk and potential transfers from stables of sixteen French farms. Int J Food Microbiol 146:253–262
Vasdinyei R, Deák T (2003) Characterization of yeast isolates originating from Hungarian dairy products using traditional and molecular identification techniques. Int J Food Microbiol 86:123–130
Viljoen BC, Knox AM, De Jager PH, Lourens-Hattingh A (2003) Development of yeast populations during processing and ripening of blue veined cheese. Food Technol Biotechnol 41:291–297
Zheng Q-Y, Westermark S-O, Rasmuson-Lestander Â, Wang X-R (2006) Detection and quantification of Cladosporium in aerosols by real-time PCR. J Environ Monit 8:153–160
Acknowledgments
This work was funded by the Novalait Inc. Fonds Québécois de la Recherche sur la Nature et les Technologies (FQRNT), the Ministère de l’Agriculture, des Pêcheries et de l’Alimentation du Québec (MAPAQ), and Agriculture and Agri-Food Canada (AAFC).
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary materials
Below is the link to the electronic supplementary material.
Online resource 1
Yeasts and molds isolated from raw cow milk from dairy farm A over a 5-month period. The concentrations of the isolates were estimated based on the proportions of each isolate and the total yeast and mold count (CFU.mL−1) in order to provide a clearer picture of the results. (DOCX 36 kb)
Online resource 2
Yeasts and molds isolated from raw cow milk from dairy farm C over a 5-month period. The concentrations of the isolates were estimated based on the proportions of each isolate and the total yeast and mold count (CFU.mL−1) in order to provide a clearer picture of the results. (DOCX 37 kb)
Online resource 3
Yeasts and molds isolated from raw cow milk from dairy farm D over a 5-month period. The concentrations of the isolates were estimated based on the proportions of each isolate and the total yeast and mold count (CFU.mL−1) in order to provide a clearer picture of the results. (DOCX 37 kb)
Online resource 4
Yeasts and molds isolated from raw cow milk from dairy farm B over a 5-month period. The concentrations of the isolates were estimated based on the proportions of each isolate and the total yeast and mold count (CFU.mL−1) in order to provide a clearer picture of the results. (DOCX 37 kb)
Rights and permissions
This article is published under an open access license. Please check the 'Copyright Information' section either on this page or in the PDF for details of this license and what re-use is permitted. If your intended use exceeds what is permitted by the license or if you are unable to locate the licence and re-use information, please contact the Rights and Permissions team.
About this article
Cite this article
Lavoie, K., Touchette, M., St-Gelais, D. et al. Characterization of the fungal microflora in raw milk and specialty cheeses of the province of Quebec. Dairy Sci. & Technol. 92, 455–468 (2012). https://doi.org/10.1007/s13594-011-0051-4
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13594-011-0051-4
Keywords
- Artisanal cheese
- Fungi
- Yeast
- Molds
- Candida krusei
- Issatchenkia orientalis
- Pichia kudriavzevii
- Multilocus sequence typing
- MLST
- Non-starter yeasts and molds