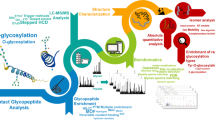
Abstract
Glycomics is the comprehensive study of glycan expression in an organism, cell, or tissue that relies on effective analytical technologies to understand glycan structure–function relationships. Owing to the macro- and micro-heterogeneity of oligosaccharides, detailed structure characterization has required an orthogonal approach, such as a combination of specific exoglycosidase digestions, LC-MS/MS, and the development of bioinformatic resources to comprehensively profile a complex biological sample. Liquid chromatography-electrospray ionization-mass spectrometry (LC-ESI-MS/MS) has emerged as a key tool in the structural analysis of oligosaccharides because of its high sensitivity, resolution, and robustness. Here, we present a strategy that uses LC-ESI-MS/MS to characterize over 200 N- and O-glycans from human saliva glycoproteins, complemented by sequential exoglycosidase treatment, to further verify the annotated glycan structures. Fragment-specific substructure diagnostic ions were collated from an extensive screen of the literature available on the detailed structural characterization of oligosaccharides and, together with other specific glycan structure feature ions derived from cross-ring and glycosidic-linkage fragmentation, were used to characterize the glycans and differentiate isomers. The availability of such annotated mass spectrometric fragmentation spectral libraries of glycan structures, together with such substructure diagnostic ions, will be key inputs for the future development of the automated elucidation of oligosaccharide structures from MS/MS data.

ᅟ


Similar content being viewed by others
References
Khoury, G.A., Baliban, R.C., Floudas, C.A.: Proteome-wide post-translational modification statistics: frequency analysis and curation of the swiss-prot database. Sci. Rep. 1(90), 1–5 (2011)
Harvey, D.J.: Fragmentation of negative ions from carbohydrates. Part 2. Fragmentation of high-mannose N-linked glycans. J. Am. Soc. Mass Spectrom. 16, 631–646 (2005)
Harvey, D.J., Dwek, R.A., Rudd, P.M.: Determining the structure of glycan moieties by mass spectrometry. Curr. Protoc. Protein Sci. 43, Unit 12.17.1–12.17.18 (2006)
Harvey, D.J., Rudd, P.M.: Fragmentation of negative ions from N-linked carbohydrates. Part 5. Anionic N-linked glycans. Int. J. Mass Spectrom 305, 120–130 (2011)
Karlsson, N.G., Schulz, B.L., Packer, N.H.: Structural determination of neutral O-linked oligosaccharide alditols by negative ion LC-electrospray-MSn. J. Am. Soc. Mass Spectrom. 15, 659–672 (2004)
Pabst, M., Altmann, F.: Glycan analysis by modern instrumental methods. Proteomics 11, 631–643 (2011)
Geyer, H., Geyer, R.: Strategies for analysis of glycoprotein glycosylation. Biochim. Biophys. Acta 1764, 1853–1869 (2006)
Bielik, A.M., Zaia, J.: Historical overview of glycoanalysis. Methods Mol. Biol. 600, 9–30 (2010)
Stumpo, K.A., Reinhold, V.N.: The N-glycome of human plasma. J. Proteome Res. 9, 4823–4830 (2010)
Prien, J.M., Prater, B.D., Cockrill, S.L.: A multi-method approach toward de novo glycan characterization: a Man-5 case study. Glycobiology 20, 629–647 (2010)
Jiao, J., Zhang, H., Reinhold, V.N.: High Performance IT-MS sequencing of glycans (spatial resolution of ovalbumin isomers). Int. J. Mass Spectrom. 303, 109–117 (2011)
Pabst, M., Bondili, J.S., Stadlmann, J., Mach, L., Altmann, F.: Mass + retention time = structure: a strategy for the analysis of N-glycans by carbon LC-ESI-MS and its application to fibrin N-glycans. Anal. Chem. 79, 5051–5057 (2007)
Royle, L., Campbell, M.P., Radcliffe, C.M., White, D.M., Harvey, D.J., Abrahams, J.L., Kim, Y.G., Henry, G.W., Shadick, N.A., Weinblatt, M.E., Lee, D.M., Rudd, P.M., Dwek, R.A.: HPLC-based analysis of serum N-glycans on a 96-well plate platform with dedicated database software. Anal. Biochem. 376, 1–12 (2008)
Rakus, J.F., Mahal, L.K.: New technologies for glycomic analysis: toward a systematic understanding of the glycome. Annu Rev Anal Chem (Palo Alto, CA) 4, 367–392 (2011)
Wacker, C., Berger, C.N., Girard, P., Meier, R.: Glycosylation profiles of therapeutic antibody pharmaceuticals. Eur. J. Pharm. Biopharm. 79, 503–507 (2011)
Joshi, H.J., Harrison, M.J., Schulz, B.L., Cooper, C.A., Packer, N.H., Karlsson, N.G.: Development of a mass fingerprinting tool for automated interpretation of oligosaccharide fragmentation data. Proteomics 4, 1650–1664 (2004)
Lohmann, K.K., von der Lieth, C.W.: GlycoFragment and GlycoSearchMS: web tools to support the interpretation of mass spectra of complex carbohydrates. Nucleic Acids Res. 32, W261–266 (2004)
Eng, J.K., Mccormack, A.L., Yates, J.R.: An approach to correlate tandem mass-spectral data of peptides with amino-acid-sequences in a protein database. J. Am. Soc. Mass Spectrom. 5, 976–989 (1994)
Maass, K., Ranzinger, R., Geyer, H., von der Lieth, C.W., Geyer, R.: “Glyco-peakfinder”—de novo composition analysis of glycoconjugates. Proteomics 7, 4435–4444 (2007)
Ceroni, A., Maass, K., Geyer, H., Geyer, R., Dell, A., Haslam, S.M.: GlycoWorkbench: a tool for the computer-assisted annotation of mass spectra of glycans. J. Proteome Res. 7, 1650–1659 (2008)
Tang, H., Mechref, Y., Novotny, M.V.: Automated interpretation of MS/MS spectra of oligosaccharides. Bioinformatics 21, 431–439 (2005)
Goldberg, D., Sutton-Smith, M., Paulson, J., Dell, A.: Automatic annotation of matrix-assisted laser desorption/ionization N-glycan spectra. Proteomics 5, 865–875 (2005)
Goldberg, D., Bern, M., Li, B.S., Lebrilla, C.B.: Automatic determination of O-glycan structure from fragmentation spectra. J. Proteome Res. 5, 1429–1434 (2006)
Hayes, C.A., Karlsson, N.G., Struwe, W.B., Lisacek, F., Rudd, P.M., Packer, N.H., Campbell, M.P.: UniCarb-DB: a database resource for glycomic discovery. Bioinformatics 27, 1343–1344 (2011)
Estrella, R.P., Whitelock, J.M., Packer, N.H., Karlsson, N.G.: The glycosylation of human synovial lubricin: implications for its role in inflammation. Biochem. J. 429, 359–367 (2010)
Issa, S., Moran, A.P., Ustinov, S.N., Lin, J.H., Ligtenberg, A.J., Karlsson, N.G.: O-linked oligosaccharides from salivary agglutinin: Helicobacter pylori binding sialyl-Lewis x and Lewis b are terminating moieties on hyperfucosylated oligo-N-acetyllactosamine. Glycobiology 20, 1046–1057 (2010)
Karlsson, N.G., Thomsson, K.A.: Salivary MUC7 is a major carrier of blood group I type O-linked oligosaccharides serving as the scaffold for sialyl Lewis x. Glycobiology 19, 288–300 (2009)
Everest-Dass, A.V., Jin, D., Thaysen-Andersen, M., Nevalainen, H., Kolarich, D., Packer, N.H.: Comparative structural analysis of the glycosylation of salivary and buccal cell proteins: innate protection against infection by C. albicans. Glycobiology 22, 1465–1479 (2012)
Guile, G.R., Harvey, D.J., O’Donnell, N., Powell, A.K., Hunter, A.P., Zamze, S., Fernandes, D.L., Dwek, R.A., Wing, D.R.: Identification of highly fucosylated N-linked oligosaccharides from the human parotid gland. Eur. J. Biochem. 258, 623–656 (1998)
Guttman, A.: Analysis of monosaccharide composition by capillary electrophoresis. J. Chromatogr. A 763, 271–277 (1997)
Drake, P.M., Cho, W., Li, B., Prakobphol, A., Johansen, E., Anderson, N.L., Regnier, F.E., Gibson, B.W., Fisher, S.J.: Sweetening the pot: adding glycosylation to the biomarker discovery equation. Clin. Chem. 56, 223–236 (2010)
Jensen, P.H., Karlsson, N.G., Kolarich, D., Packer, N.H.: Structural analysis of N- and O-glycans released from glycoproteins. Nat. Protoc. 7, 1299–1310 (2012)
Harvey, D.J.: Fragmentation of negative ions from carbohydrates. Part 3. Fragmentation of hybrid and complex N-linked glycans. J. Am. Soc. Mass Spectrom. 16, 647–659 (2005)
Harvey, D.J., Royle, L., Radcliffe, C.M., Rudd, P.M., Dwek, R.A.: Structural and quantitative analysis of N-linked glycans by matrix-assisted laser desorption ionization and negative ion nanospray mass spectrometry. Anal. Biochem. 376, 44–60 (2008)
Robbe, C., Michalski, J.C., Capon, C.: Structural determination of O-glycans by tandem mass spectrometry. Methods Mol. Biol. 347, 109–123 (2006)
Chai, W., Piskarev, V.E., Mulloy, B., Liu, Y., Evans, P.G., Osborn, H.M., Lawson, A.M.: Analysis of chain and blood group type and branching pattern of sialylated oligosaccharides by negative ion electrospray tandem mass spectrometry. Anal. Chem. 78, 1581–1592 (2006)
Deguchi, K., Takegawa, Y., Ito, H., Miura, N., Yoshioka, S., Nagai, S., Nakagawa, H., Nishimura, S.: Structural assignment of isomeric 2-aminopyridine-derivatized monosialylated biantennary N-linked oligosaccharides using negative-ion multistage tandem mass spectral matching. Rapid Commun. Mass Spectrom. 20, 412–418 (2006)
Domon, B., Costello, C.E.: A systematic nomenclature for carbohydrate fragmentations in FAB-MS/MS spectra of glycoconjugates. Glycoconj. J. 5, 397–409 (1988)
Cooper, C.A., Gasteiger, E., Packer, N.H.: GlycoMod—a software tool for determining glycosylation compositions from mass spectrometric data. Proteomics 1, 340–349 (2001)
Plasencia, M.D., Isailovic, D., Merenbloom, S.I., Mechref, Y., Novotny, M.V., Clemmer, D.E.: Resolving and assigning N-linked glycan structural isomers from ovalbumin by IMS-MS. J. Am. Soc. Mass Spectrom. 19, 1706–1715 (2008)
Ito, H., Yamada, K., Deguchi, K., Nakagawa, H., Nishimura, S.: Structural assignment of disialylated biantennary N-glycan isomers derivatized with 2-aminopyridine using negative-ion multistage tandem mass spectral matching. Rapid Commun. Mass Spectrom. 21, 212–218 (2007)
Prien, J.M., Huysentruyt, L.C., Ashline, D.J., Lapadula, A.J., Seyfried, T.N., Reinhold, V.N.: Differentiating N-linked glycan structural isomers in metastatic and nonmetastatic tumor cells using sequential mass spectrometry. Glycobiology 18, 353–366 (2008)
Seymour, J.L., Costello, C.E., Zaia, J.: The influence of sialylation on glycan negative ion dissociation and energetics. J. Am. Soc. Mass Spectrom. 17, 844–854 (2006)
Sagi, D., Peter-Katalinic, J., Conradt, H.S., Nimtz, M.: Sequencing of tri- and tetraantennary N-glycans containing sialic acid by negative mode ESI QTOF tandem MS. J. Am. Soc. Mass Spectrom. 13, 1138–1148 (2002)
Mechref, Y., Kang, P., Novotny, M.V.: Differentiating structural isomers of sialylated glycans by matrix-assisted laser desorption/ionization time-of-flight/time-of-flight tandem mass spectrometry. Rapid Commun. Mass Spectrom. 20, 1381–1389 (2006)
Zaia, J.: Mass spectrometry of oligosaccharides. Mass Spectrom. Rev. 23, 161–227 (2004)
Wheeler, S.F., Harvey, D.J.: Negative ion mass spectrometry of sialylated carbohydrates: discrimination of N-acetylneuraminic acid linkages by MALDI-TOF and ESI-TOF mass spectrometry. Anal. Chem. 72, 5027–5039 (2000)
Nakano, M., Saldanha, R., Gobel, A., Kavallaris, M., Packer, N.H.: Identification of glycan structure alterations on cell membrane proteins in desoxyepothilone B resistant leukemia cells. Mol. Cell. Proteom. 10(M111), 009001–009012 (2011)
Brockhausen, I., Schachter, H., Stanley, P.: In Essentials of Glycobiology. Varki, A., Cummings, R.D., Esko, J.D., Freeze, H.H., Stanley, P., Certozzi, C.R., Hart, G.W., Etzler, M.E., Eds., Cold Spring Harbor, Cold Spring Harbor Laboratory Press, pp. 115–128 (2009)
Robbe, C., Capon, C., Coddeville, B., Michalski, J.C.: Diagnostic ions for the rapid analysis by nano-electrospray ionization quadrupole time-of-flight mass spectrometry of O-glycans from human mucins. Rapid Commun. Mass Spectrom. 18, 412–420 (2004)
Kolarich, D., Weber, A., Turecek, P.L., Schwarz, H.P., Altmann, F.: Comprehensive glyco-proteomic analysis of human α1-antitrypsin and its charge isoforms. Proteomics 6, 3369–3380 (2006)
Ruhaak, L.R., Miyamoto, S., Kelly, K., Lebrilla, C.B.: N-Glycan profiling of dried blood spots. Anal. Chem. 84, 396–402 (2012)
Hua, S., An, H.J., Ozcan, S., Ro, G.S., Soares, S., DeVere-White, R., Lebrilla, C.B.: Comprehensive native glycan profiling with isomer separation and quantitation for the discovery of cancer biomarkers. Analyst 136, 3663–3671 (2011)
Varki, A., Cummings, R.D., Esko, J.D., Freeze, H.H., Stanley, P., Marth, J.D., Bertozzi, C.R., Hart, G.W., Etzler, M.E.: Symbol nomenclature for glycan representation. Proteomics 9, 5398–5399 (2009)
Harvey, D.J., Merry, A.H., Royle, L., Campbell, M.P., Rudd, P.M.: Symbol nomenclature for representing glycan structures: extension to cover different carbohydrate types. Proteomics 11, 4291–4295 (2011)
Acknowledgment
The authors acknowledge Dr. Niclas Karlsson and Dr. Catherine Hayes for their ongoing collaboration with the UniCarb-DB database project. This research project was facilitated by access to the Australian Proteomics Analysis Facility (APAF) established under the Australian Government NCRIS program.
This work was funded by Macquarie University Research Excellence Scheme postgraduate scholarship and Cystic Fibrosis Australia postgraduate studentship grant, Australian National Data Service (ANDS) grant DC12A and National eResearch Collaboration Tools and Resources (NeCTAR) grant RT016.
Author information
Authors and Affiliations
Corresponding author
Electronic Supplementary Material
Figure S1
Workflow for glycan analysis including bioinformatic support for the annotation and interpretation of the generated MS/MS spectra. N- and O-linked glycans were released sequentially from salivary proteins dot blotted on to PVDF membranes, by PNGase F and reductive β-elimination and subsequently analyzed by PGC LC-ESI-IT MS/MS (PNG 158 kb)
Figure S2
Identification and confirmation of sialic acid linkage and arm location of monosialyated biantennary N-glycan isomers. MS/MS spectra were obtained from the doubly charged negative ion parent ions of m/z 965.82– (NeuAc1Hex5HexNAc4) eluting at 44.5 min (a), 45.5 min (b), 52.5 min (c), and 53.5 min (d); (e) shows the extracted ion chromatogram of the four m/z 965.82- isomers (JPEG 243 kb)
Figure S3
Identification and characterization of sialylated O-glycan isomers. (a) and (b) The negative ion fragmentation spectra of m/z 878.51– (NeuAc1Hex1HexNAc2) isomers eluting at 25 and 26.7 min, respectively. The extracted ion chromatogram of the m/z 878.52– isomers is shown in (c) (JPEG 86 kb)
Figure S4
Identification and characterization of sulphated O-glycan isomers. Negative ion fragmentation spectra of m/z 8131– (Sulph1Hex1HexNAc2) isomers eluting at 21.8 min (a) and 22.5 min (b) are shown; (c) shows an extracted ion chromatogram of m/z 8131– isomers (JPEG 90 kb)
Figure S5
Extracted ion chromatograms of m/z 1039.42– (Hex5HexNAc4dHex3) N-glycan isomers. The glycan structural and linkage isomers were orthogonally confirmed by exoglycosidase treatments. ABS: Arthrobacter ureafaciens sialidase (α2–3,6,8). BTG: bovine testes β-galactosidase (β1–3,4,6). AMF: Almond meal α-fucosidase (α1–3,4). BKF: bovine kidney α-fucosidase (α1–2, 6) (JPEG 64 kb)
Figure S6
Glycan annotation report generated by GlycoWorkBench tool [20] of monosialyated biantennary N-glycan isomers of m/z 965.82– (NeuAc1Hex5HexNAc4) differing in α2-6 sialylation of the antennae. (a) MS/MS spectra of the doubly charged negative ion eluting at 44.5 min and its GlycoWorkBench generated annotation based on in silico fragmentation (b) MS/MS spectra of the doubly charged negative ion eluting at 45.5 min and its Glycoworkbench generated annotation based on in silico fragmentation. To simplify the figure, only singly charged glycan fragment masses above a 3 % intensity threshold are displayed. (c) Statistical coverage from the two isomers generated by Glycoworkbench (JPEG 207 kb)
Rights and permissions
About this article
Cite this article
Everest-Dass, A.V., Abrahams, J.L., Kolarich, D. et al. Structural Feature Ions for Distinguishing N- and O-Linked Glycan Isomers by LC-ESI-IT MS/MS. J. Am. Soc. Mass Spectrom. 24, 895–906 (2013). https://doi.org/10.1007/s13361-013-0610-4
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13361-013-0610-4