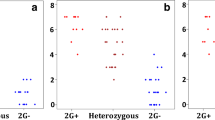
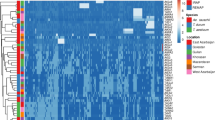
Abstract
Variability of the beta-amylase gene in bread wheat, artificial amphidiploids, and derived introgression wheat lines was analyzed. Variation in homeologous beta-amylase sequences caused by the presence of MITE (Miniature Inverted-Repeat Transposable Element) and its footprint has been identified in bread wheat. The previously unknown location of MITE in Triticum urartu and T. aestivum L. beta-amylase gene has been found. These species have a MITE sequence in the third intron of beta-amylase, as opposed to Aegilops comosa and a number of other Triticeae species, which have it in the fourth intron. These two MITEs from Ae. comosa and T. aestivum were shown to have low identity scores. Miosa, an artificial amphidiploid, which has the M genome from Ae. comosa was shown to lose the MITE sequences. This loss might be caused by genomic shock due to allopolyploidization.







Similar content being viewed by others
References
Antonyuk MZ, Bodylyova MV, Ternovskaya TK (2009) Genome structure of introgressive lines Triticum aestivum/Aegilops sharonensis. Cytol Genet 43:58–67
Antonyuk MZ, Shpylchyn VV, Ternovska TK (2013) Permanent genetic variability in the introgressive lines and amphidiploids of Triticeae. Cytol Genet 47:58–68
Bureau TE, Wessler SR (1994) Stowaway: a new family of inverted repeat elements associated with the genes of both monocotyledonous and dicotyledonous plants. Plant Cell 6:907–916
Chen L, Jiongjiong C, Yu Z, Qun H, Wenqing S, Hanhui K (2012) Miniature Inverted–Repeat Transposable Elements (MITEs) have been accumulated through amplification bursts and play important roles in gene expression and species diversity in Oryza sativa. Mol Biol Evol 29:1005–1017
Endo TR (2007) The gametocidal chromosome as a tool for chromosome manipulation in wheat. Chromosome Res 15:67–75. doi:10.1007/s10577-006-1100-3
Feldman M, Levy A (2009) Genome evolution in allopolyploid wheat — a revolutionary reprogramming followed by gradual changes. J Genet Genom 36:511–518. doi:10.1016/S1673-8527(08)60142-3
Feldman M, Sears ER (1981) The wild gene resources of wheat. Sci Am 244:102–112
Guermonprez H, Henaff E, Cifuentes M, Casacuberta JM (2012) MITEs, miniature elements with a major role in plant genome evolution. In: Grandbastien MA, Casacuberta JM (eds) Plant transposable elements: impact on genome structure and function. Springer, Berlin, pp 113–124
Han FP, Fedak G, Ouellet T, Liu B (2003) Rapid genomic changes in interspecific and intergeneric hybrids and allopolyploids of Triticeae. Genome 46:716–723. doi:10.1139/G03-049
Han FP, Liu B, Fedak G, Liu ZH (2004) Genomic constitution and variation in five partial amphiploids of wheat — Thinopyrum intermedium as revealed by GISH, multicolor GISH and seed storage protein analysis. Theor Appl Genet 109:1070–1076
Han FP, Fedak G, Guo W, Liu B (2005) Rapid and repeatable elimination of a parental genome-specific DNA repeat (pGc1R-1a) in newly synthesized wheat allopolyploids. Genetics 170:1239–1245. doi:10.1534/genetics.104.039263
Jackson S, Chen ZJ (2010) Genomic and expression plasticity of polyploidy. Curr Opin Plant Biol 13:153–159. doi:10.1016/j.pbi.2009.11.004
Korbie DJ, Mattick JS (2008) Touchdown PCR for increased specificity and sensitivity in PCR amplification. Nat Protoc 3:1452–1456. doi:10.1038/nprot.2008.133
Kuang H, Padmanabhan C, Li F, Kamei A, Bhaskar PB, Ouyang S, Jiang J, Buell CR, Baker B (2009) Identification of miniature inverted-repeat transposable elements (MITEs) and biogenesis of their siRNAs in the Solanaceae: New functional implications for MITEs. Genome Res 19:42–56
Liu B, Xu C, Zhao N, Qi B, Kimatu JN, Pang J, Han F (2009) Rapid genomic changes in polyploid wheat and related species: implications for genome evolution and genetic improvement. J Genet Genom 36:519–528. doi:10.1016/S1673-8527(08)60143-5
Ma X-F, Gustafson JP (2008) Allopolyploidization-accommodated genomic sequence changes in Triticale. Ann Bot 101:825–832. doi:10.1093/aob/mcm331
Mason-Gamer RJ (2005) The beta-amylase genes of grasses and a phylogenetic analysis of the Triciceae (Poaceae). Am J Bot 92:1045–1058
Mason-Gamer RJ (2007) Multiple homoplasious insertions and deletions of a Triticeae (Poaceae) DNA transposon: a phylogenetic perspective. BMC Evol Biol 7:92. doi:10.1186/1471-2148-7-92
Masterson J (1994) Stomatal size in fossil plants: evidence for polyploidy in majority of angiosperms. Science 264:421–424
Nasuda S, Friebe B, Gill BS (1998) Gametocidal genes induce chromosome breakage in the interphase prior to the first mitotic cell division of the male gametophyte in wheat. Genetics 149:1115–1124
Navalikhina AG, Antonyuk MZ, Ternovska TK (2014) Genetic variability of introgressive wheat lines by beta-amylase gene. Faktory eksperymental’noyi evolyutsiyi orhanizmiv 14:67–71 (in Ukrainian)
Olivera PD, Steffenson BJ (2009) Aegilops sharonensis: origin, genetics, diversity, and potential for wheat improvement. Botany 87:740–756
Predict a Secondary Structure Web Server. Retrieved from http://rna.urmc.rochester.edu/RNAstructureWeb/index.html
Sambrook J, Fritsch EF, Maniatis T (1989) Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, New York
Soltis DE, Albert VA, Leebens-Mack J, Bell CD, Paterson AH, Zheng C, Sankoff D, dePamphilis CW, Wall KP, Soltis PS (2009) Polyploidy and angiosperm diversification. Am J Bot 96:336–348
Wicker T, Sabot F, Hua-Van A, Bennetzen JL, Capy P, Chalhoub B, Flavell A, Leroy P, Morgante M, Panaud O, Paux E, SanMiguel P, Schulman AH (2007) A unified classification system for eukaryotic transposable elements. Nat Rev Genet 8:973–982
Zhirov EG, Ternovskaya TK (1984) Genome engineering in wheat. Vestnik S-kh Nauk 10:58–66 (in Russian)
Acknowledgments
We are pleased to thank George Fedak (Ottawa, Canada) for helpful remarks and careful review.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
Maksym Antonyuk declares that he has no conflict of interest.
Anastasiia Navalikhina declares that she has no conflict of interest.
Tamara Ternovska declares that she has no conflict of interest.
Human and animal rights and informed consent
This article does not contain any studies with human participants or animals performed by any of the authors.
Additional information
Communicated by: Andrzej Górny
Rights and permissions
About this article
Cite this article
Antonyuk, M., Navalikhina, A. & Ternovska, T. Beta-amylase gene variability in introgressive wheat lines. J Appl Genetics 58, 143–149 (2017). https://doi.org/10.1007/s13353-016-0364-3
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13353-016-0364-3