Abstract
Human epithelial cadherin (E-cadherin), a member of transmembrane glycoprotein family, encoded by the E-cadherin gene, plays a key role in cell-cell adhesion, adherent junction in normal epithelial tissues, contributing to tissue differentiation and homeostasis. Although previous studies indicated that inactivation of the E-cadherin is mainly induced by hypermethylation of E-cadherin gene, evidence concerning E-cadherin hypermethylation in the carcinogenesis and development of non-small cell lung carcinoma (NSCLC) remains controversial. In this study, we conducted a meta-analysis to quantitatively evaluate the effects of E-cadherin hypermethylation on the incidence and clinicopathological characteristics of NSCLC. A comprehensive search of PubMed and Embase databases was performed up to October 2014. Analyses of pooled data were performed. Odds ratios (ORs) were calculated and summarized. Our meta-analysis combining 18 published articles demonstrated that the hypermethylation frequencies in NSCLC were significantly higher than those in normal control tissues, OR = 3.55, 95 % confidence interval (CI) = 1.98–6.36, p < 0.0001. Further analysis showed that E-cadherin hypermethylation was not strongly associated with the sex or smoking status in NSCLC patients. In addition, E-cadherin hypermethylation was also not strongly associated with pathological types, differentiated status, clinical stages, or metastatic status in NSCLC patients. The results from the current study indicate that the hypermethylation frequency of E-cadherin in NSCLC is strongly associated with NSCLC incidence and it may be an early event in carcinogenesis of NSCLC. We also discussed the potential value of E-cadherin as a drug target that may bring new direction and hope for cancer treatment through gene-targeted therapy.







Similar content being viewed by others
References
Jemal A, Bray F, Center MM, Ferlay J, Ward E, et al. Global cancer statistics. CA Cancer J Clin. 2011;61:69–90.
Ramalingam S, Belani C. Systemic chemotherapy for advanced non-small cell lung cancer: recent advances and future directions. Oncologist. 2008;13 Suppl 1:5–13.
Ghavifekr Fakhr M, Farshdousti Hagh M, Shanehbandi D, Baradaran B. DNA methylation pattern as important epigenetic criterion in cancer. Genet Res Int. 2013;2013:317569.
Delpu Y, Cordelier P, Cho WC, Torrisani J. DNA methylation and cancer diagnosis. Int J Mol Sci. 2013;14:15029–58.
Ma X, Wang YW, Zhang MQ, Gazdar AF. DNA methylation data analysis and its application to cancer research. Epigenomics. 2013;5:301–16.
Walter K, Holcomb T, Januario T, Du P, Evangelista M, et al. DNA methylation profiling defines clinically relevant biological subsets of non-small cell lung cancer. Clin Cancer Res. 2012;18:2360–73.
Fleischhacker M, Dietrich D, Liebenberg V, Field JK, Schmidt B. The role of DNA methylation as biomarkers in the clinical management of lung cancer. Expert Rev Respir Med. 2013;7:363–83.
Bussemakers MJ, van Bokhoven A, Mees SG, Kemler R, Schalken JA. Molecular cloning and characterization of the human E-cadherin cDNA. Mol Biol Rep. 1993;17:123–8.
Rodriguez FJ, Lewis-Tuffin LJ, Anastasiadis PZ. E-cadherin’s dark side: possible role in tumor progression. Biochim Biophys Acta. 2012;1826:23–31.
van RF. Beyond E-cadherin: roles of other cadherin superfamily members in cancer. Nat Rev Cancer. 2014;14:121–34.
Karayiannakis AJ, Syrigos KN, Chatzigianni E, Papanikolaou S, Alexiou D, et al. Aberrant E-cadherin expression associated with loss of differentiation and advanced stage in human pancreatic cancer. Anticancer Res. 1998;18:4177–80.
Zheng Z, Pan J, Chu B, Wong YC, Cheung AL, et al. Downregulation and abnormal expression of E-cadherin and beta-catenin in nasopharyngeal carcinoma: close association with advanced disease stage and lymph node metastasis. Hum Pathol. 1999;30:458–66.
Oka H, Shiozaki H, Kobayashi K, Inoue M, Tahara H, et al. Expression of E-cadherin cell adhesion molecules in human breast cancer tissues and its relationship to metastasis. Cancer Res. 1993;53:1696–701.
Su CY, Li YS, Han Y, Zhou SJ, Liu ZD. Correlation between expression of cell adhesion molecules CD44 v6 and E-cadherin and lymphatic metastasis in non-small cell lung cancer. Asian Pac J Cancer Prev. 2014;15:2221–4.
Ponce E, Louie MC, Sevigny MB. Acute and chronic cadmium exposure promotes E-cadherin degradation in MCF7 breast cancer cells. Mol Carcinog. 2014.
Schildberg CW, Abba M, Merkel S, Agaimy A, Dimmler A, et al. Gastric cancer patients less than 50 years of age exhibit significant downregulation of E-cadherin and CDX2 compared to older reference populations. Adv Med Sci. 2014;59:142–6.
Altman DG, McShane LM, Sauerbrei W, Taube SE. Reporting recommendations for tumor marker prognostic studies (REMARK): explanation and elaboration. BMC Med. 2012;10:51.
Sculier JP, Ghisdal L, Berghmans T, Branle F, Lafitte JJ, et al. The role of mitomycin in the treatment of non-small cell lung cancer: a systematic review with meta-analysis of the literature. Br J Cancer. 2001;84:1150–5.
Huedo-Medina TB, Sanchez-Meca J, Marin-Martinez F, Botella J. Assessing heterogeneity in meta-analysis: Q statistic or I2 index? Psychol Methods. 2006;11:193–206.
Higgins JP, Thompson SG, Deeks JJ, Altman DG. Measuring inconsistency in meta-analyses. BMJ. 2003;327:557–60.
DerSimonian R. Meta-analysis in the design and monitoring of clinical trials. Stat Med. 1996;15:1237–48. discussion 1249–1252.
Begg CB, Mazumdar M. Operating characteristics of a rank correlation test for publication bias. Biometrics. 1994;50:1088–101.
Zheng SY, Hou JY, Zhao J, Jiang D, Ge JF, et al. Clinical outcomes of downregulation of E-cadherin gene expression in non-small cell lung cancer. Asian Pac J Cancer Prev. 2012;13:1557–61.
Guzman L, Depix MS, Salinas AM, Roldan R, Aguayo F, et al. Analysis of aberrant methylation on promoter sequences of tumor suppressor genes and total DNA in sputum samples: a promising tool for early detection of COPD and lung cancer in smokers. Diagn Pathol. 2012;7:87.
Begum S, Brait M, Dasgupta S, Ostrow KL, Zahurak M, et al. An epigenetic marker panel for detection of lung cancer using cell-free serum DNA. Clin Cancer Res. 2011;17:4494–503.
Sasaki H, Hikosaka Y, Kawano O, Moriyama S, Yano M, et al. Methylation of the DLEC1 gene correlates with poor prognosis in Japanese lung cancer patients. Oncol Lett. 2010;1:283–7.
Vaissiere T, Hung RJ, Zaridze D, Moukeria A, Cuenin C, et al. Quantitative analysis of DNA methylation profiles in lung cancer identifies aberrant DNA methylation of specific genes and its association with gender and cancer risk factors. Cancer Res. 2009;69:243–52.
Buckingham L, Penfield FL, Kim A, Liptay M, Barger C, et al. PTEN, RASSF1 and DAPK site-specific hypermethylation and outcome in surgically treated stage I and II nonsmall cell lung cancer patients. Int J Cancer. 2010;126:1630–9.
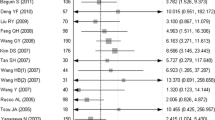
Wang G, Hu X, Lu C, Su C, Luo S, et al. Promoter-hypermethylation associated defective expression of E-cadherin in primary non-small cell lung cancer. Lung Cancer. 2008;62:162–72.
Feng Q, Hawes SE, Stern JE, Wiens L, Lu H, et al. DNA methylation in tumor and matched normal tissues from non-small cell lung cancer patients. Cancer Epidemiol Biomarkers Prev. 2008;17:645–54.
Wang Y, Zhang D, Zheng W, Luo J, Bai Y, et al. Multiple gene methylation of nonsmall cell lung cancers evaluated with 3-dimensional microarray. Cancer. 2008;112:1325–36.
Tan SH, Ida H, Lau QC, Goh BC, Chieng WS, et al. Detection of promoter hypermethylation in serum samples of cancer patients by methylation-specific polymerase chain reaction for tumour suppressor genes including RUNX3. Oncol Rep. 2007;18:1225–30.
Kim DS, Kim MJ, Lee JY, Kim YZ, Kim EJ, et al. Aberrant methylation of E-cadherin and H-cadherin genes in nonsmall cell lung cancer and its relation to clinicopathologic features. Cancer. 2007;110:2785–92.
Gu J, Berman D, Lu C, Wistuba II, Roth JA, et al. Aberrant promoter methylation profile and association with survival in patients with non-small cell lung cancer. Clin Cancer Res. 2006;12:7329–38.
Nakata S, Sugio K, Uramoto H, Oyama T, Hanagiri T, et al. The methylation status and protein expression of CDH1, p16(INK4A), and fragile histidine triad in nonsmall cell lung carcinoma: epigenetic silencing, clinical features, and prognostic significance. Cancer. 2006;106:2190–9.
Tsou JA, Shen LY, Siegmund KD, Long TI, Laird PW, et al. Distinct DNA methylation profiles in malignant mesothelioma, lung adenocarcinoma, and non-tumor lung. Lung Cancer. 2005;47:193–204.
Russo AL, Thiagalingam A, Pan H, Califano J, Cheng KH, et al. Differential DNA hypermethylation of critical genes mediates the stage-specific tobacco smoke-induced neoplastic progression of lung cancer. Clin Cancer Res. 2005;11:2466–70.
Topaloglu O, Hoque MO, Tokumaru Y, Lee J, Ratovitski E, et al. Detection of promoter hypermethylation of multiple genes in the tumor and bronchoalveolar lavage of patients with lung cancer. Clin Cancer Res. 2004;10:2284–8.
Yanagawa N, Tamura G, Oizumi H, Takahashi N, Shimazaki Y, et al. Promoter hypermethylation of tumor suppressor and tumor-related genes in non-small cell lung cancers. Cancer Sci. 2003;94:589–92.
Zochbauer-Muller S, Fong KM, Virmani AK, Geradts J, Gazdar AF, et al. Aberrant promoter methylation of multiple genes in non-small cell lung cancers. Cancer Res. 2001;61:249–55.
Akhavan-Niaki H, Samadani AA. DNA methylation and cancer development: molecular mechanism. Cell Biochem Biophys. 2013;67:501–13.
Ponce E, Louie MC, Sevigny MB. Acute and chronic cadmium exposure promotes E-cadherin degradation in MCF7 breast cancer cells. Mol Carcinog. 2014;5:22170.
Chang Z, Zhou H, Liu Y. Promoter methylation and polymorphism of E-cadherin gene may confer a risk to prostate cancer: a meta-analysis based on 22 studies. Tumour Biol. 2014;24:24.
Wu X, Zhuang YX, Hong CQ, Chen JY, You YJ, et al. Clinical importance and therapeutic implication of E-cadherin gene methylation in human ovarian cancer. Med Oncol. 2014;31:100.
Ceteci F, Ceteci S, Zanucco E, Thakur C, Becker M, et al. E-cadherin controls bronchiolar progenitor cells and onset of preneoplastic lesions in mice. Neoplasia. 2012;14:1164–77.
Liu Y, Hong Y, Zhao Y, Ismail TM, Wong Y, et al. Histone H3 (lys-9) deacetylation is associated with transcriptional silencing of E-cadherin in colorectal cancer cell lines. Cancer Investig. 2008;26:575–82.
Ling ZQ, Li P, Ge MH, Zhao X, Hu FJ, et al. Hypermethylation-modulated down-regulation of CDH1 expression contributes to the progression of esophageal cancer. Int J Mol Med. 2011;27:625–35.
Patra A, Deb M, Dahiya R, Patra SK. 5-Aza-2'-deoxycytidine stress response and apoptosis in prostate cancer. Clin Epigenetics. 2011;2:339–48.
Liu YY, Han JY, Lin SC, Liu ZY, Jiang WT. Effect of CDH1 gene methylation on transforming growth factor (TGF-beta)-induced epithelial-mesenchymal transition in alveolar epithelial cell line A549. Genet Mol Res. 2014;13.
Lopes N, Carvalho J, Duraes C, Sousa B, Gomes M, et al. 1Alpha,25-dihydroxyvitamin D3 induces de novo E-cadherin expression in triple-negative breast cancer cells by CDH1-promoter demethylation. Anticancer Res. 2012;32:249–57.
Jiang L, Chan JY, Fung KP. Epigenetic loss of CDH1 correlates with multidrug resistance in human hepatocellular carcinoma cells. Biochem Biophys Res Commun. 2012;422:739–44.
Yi TZ, Li J, Han X, Guo J, Qu Q, et al. DNMT inhibitors and HDAC inhibitors regulate E-cadherin and Bcl-2 expression in endometrial carcinoma in vitro and in vivo. Chemotherapy. 2012;58:19–29.
Sahai E, Marshall CJ. RHO-GTPases and cancer. Nat Rev Cancer. 2002;2:133–42.
Vincan E, Barker N. The upstream components of the Wnt signalling pathway in the dynamic EMT and MET associated with colorectal cancer progression. Clin Exp Metastasis. 2008;25:657–63.
Conflicts of interest
None
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Zhong, K., Chen, W., Xiao, N. et al. The clinicopathological significance and potential drug target of E-cadherin in NSCLC. Tumor Biol. 36, 6139–6148 (2015). https://doi.org/10.1007/s13277-015-3298-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13277-015-3298-1