Abstract
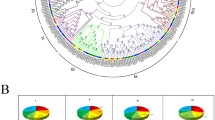
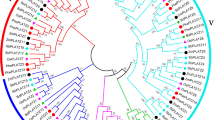
The Dof (DNA binding with One Finger) family of single zinc finger proteins is a family of plant-specific transcription factors. These transcription factors have a variety of important functions in different biological processes in plants. In the current study, we identified 26 Dof family genes in moso bamboo (Phyllostachys heterocycla var. pubescens). A complete overview of PhDof genes in moso bamboo is presented, including the gene structures, phylogeny, protein motifs and expression patterns. Phylogenetic analysis of the 26 PhDof proteins identified four classes constituting seven clusters (A, B1, C1, C2, D1, D2 and D3). In addition, a comparative analysis between the Dof genes in moso bamboo, Arabidopsis (Arabidopsis thaliana L.) and rice (Oryza sativa L.) was also performed, and several putative paralogous and orthologous genes were identified. The exon numbers in Dof genes ranged from one to three in many plants; however, the exon number in PhDofs ranged from one to four. The PhDof genes displayed differential expression in different parts of the shoot and at different flower development stages. This study represents the first step towards a genome-wide analysis of the Dof genes in moso bamboo. Our study provides a useful reference for cloning and functional analysis of members of the Dof gene family in moso bamboo and other species.





Similar content being viewed by others
References
Baumann K, De Paolis A, Costantino P, Gualberti G (1999) The DNA binding site of the Dof protein NtBBF1 is essential for tissue-specific and auxin-regulated expression of the rolB oncogene in plants. Plant Cell 11:323–333
Cai X, Zhang Y, Zhang C, Zhang T, Hu T, Ye J, Zhang J, Wang T, Li H, Ye Z (2013) Genome-wide analysis of plant-specific Dof transcription factor family in tomato. J Integr Plant Biol 6:552–566
Chen X, Wang D, Liu C, Wang M, Wang T, Zhao Q, Yu J (2012) Maize transcription factor Zmdof1 involves in the regulation of Zm401 gene. Plant Growth Regul 66:271–284
Cominelli E, Galbiati M, Albertini A, Fornara F, Conti L, Coupland G, Tonelli C (2011) DOF-binding sites additively contribute to guard cell-specificity of AtMYB60 promoter. BMC Plant Biol 11:162
de Dios Barajas-López J, Tezycka J, Travaglia CN, Serrato AJ, Chueca A, Thormählen I, Geigenberger P, Sahrawy M (2012) Expression of the chloroplast thioredoxins f and m is linked to shortterm changes in the sugar and thiol status in leaves of Pisum sativum. J Exp Bot 13:4887–4990
Deng W, Wang Y, Liu Z, Cheng H, Xue Y (2014) HemI: a toolkit for illustrating heatmaps. PLoS one 11:e111988
Diaz I, Vicente-Carbajosa J, Abraham Z, Martinez M, Moneda II, Carbonero P (2002) The GAMYB protein from barley interacts with the DOF transcription factor BPBF and activates endosperm-specific genes during seed development. Plant J 4:453–464
Dong G, Ni Z, Yao Y, Sun XN, Qi X (2007) Wheat Dof transcription factor WPBF interacts with TaQM and activates transcription of an alpha-gliadin gene during wheat seed development. Plant Mol Biol 63:73–84
Finn RD, Clements J, Eddy SR (2011) HMMER web server: interactive sequence similarity searching. Nucleic Acids Res 39:29–37
Fornara F, Panigrahi KCS, Gissot L, Sauerbrunn N, Ruhl M, Jarillo JA, Coupland G (2009) Arabidopsis DOF transcription factors act redundantly to reduce CONSTANS expression and are essential for a photoperiodic flowering response. Dev Cell 17:75–86
Gabriele S, Rizza A, Martone J, Circelli P, Costantino P, Vittorioso P (2010) The Dof protein DAG1 mediates PIL5 activity on seed germination by negatively regulating GA biosynthetic gene AtGA3ox1. Plant J 61:312–323
Gao X (2011) Study on the components and biological activity of bamboo shoot shell extractions. Dissertation, Beijing Forestry University
Gao J, Zhang Y, Zhang CL, Qi FY, Li XP, Mu SH, Peng ZH (2014) Characterization of the floral transcriptome of moso bamboo (Phyllostachys edulis) at different flowering developmental stages by transcriptome sequencing and RNA-Seq analysis. PLoS one 9:e98910
Gardiner J, Sherr I, Scarpella E (2010) Expression of DOF genes identifies early stages of vascular development in Arabidopsis leaves. Int J Dev Biol 54:1389–1396
Gaur VS, Singh US, Kumar A (2011) Transcriptional profiling and in silico analysis of Dof transcription factor gene family for understanding their regulation during seed development of rice (Oryza sativa L.). Mol Biol Rep 38:2827–2848
Gualberti G, Papi M, Bellucci L, Ricci I, Bouche D, Camilleri C, Costantino P, Vittorioso P (2002) Mutations in the Dof zinc finger genes DAG2 and DAG1 influence with opposite effects the germination of Arabidopsis seeds. Plant Cell 14:1253–1263
Guo Y, Qiu L (2013) Genome-wide analysis of the Dof transcription factor gene family reveals soybean-specific duplicable and functional characteristics. PLoS one 9:e76809
Guo Y, Qin G, Gu H, Qu L (2009) Dof5.6/HCA2, a Dof transcription factor gene, regulates interfascicular cambium formation and vascular tissue development in Arabidopsis. Plant Cell 21:3518–3534
He L, Su C, Wang Y, Wei Z (2015) ATDOF5.8 protein is the upstream regulator of ANAC069 and is responsive to abiotic stress. Biochimie 110:17–24
Hernando-Amado S, González-Calle V, Carbonero P, Barrero-Sicilia C (2012) The family of DOF transcription factors in Brachypodium distachyon: phylogenetic comparison with rice and barley DOFs and expression profiling. BMC Plant Biol 12:202
Imaizumi T, Schultz TF, Harmon FG, Ho LA, Kay SA (2005) FKF1 F-box protein mediates cyclic degradation of a repressor of CONSTANS in Arabidopsis. Science 309:293
Iwamoto M, Higo K, Takano M (2009) Circadian clock- and phytochrome-regulated Dof-like gene, Rdd1, is associated with grain size in rice. Plant Cell Environ 32:592–603
Jiang Y, Zeng B, Zhao H, Zhang M, Xie S, Lai J (2012) Genome-wide transcription factor gene prediction and their expressional tissue-specificities in maize. J Integr Plant Biol 9:616–630
Jin J, Zhang H, Kong L, Gao G, Luo J (2014a) PlantTFDB 3.0: a portal for the functional and evolutionary study of plant transcription factors. Nucleic Acids Res 42:D1182–D1187
Jin Z, Chandrasekaran U, Liu A (2014b) Genome-wide analysis of the Dof transcription factors in castor bean (Ricinus communis L.). Genes Genom 36:527–537
Kang H, Singh KB (2000) Characterization of salicylic acid-responsive, Arabidopsis Dof domain proteins: overexpression of OBP3 leads to growth defects. Plant J 4:329–339
Kang H, Foley RC, Onate-Sanchez L, Lin C, Singh KB (2003) Target genes for OBP3, a Dof transcription factor, include novel basic helix-loop-helix domain proteins inducible by salicylic acid. Plant J 35:362–372
Kawakatsu T, Takaiwa F (2010) Differences in transcriptional regulatory mechanisms functioning for free lysine content and seed storage protein accumulation in rice grain. Plant Cell Physiol 12:1964–1974
Kisu Y, Ono T, Shimofurutani N, Suzuki M, Esaka M (1998) Characterization and expression of a new class of zinc finger protein that binds to silencer region of ascorbate oxidase gene. Plant Cell Physiol 10:1054–1064
Konishi M, Yanagisawa S (2007) Sequential activation of two Dof transcription factor gene promoters during vascular development in Arabidopsis thaliana. Plant Physiol and Biochem 45:623–629
Kumar R, Taware R, Gaur VS, Guru SK, Kumar A (2009) Influence of nitrogen on the expression of TaDof1 transcription factor in wheat and its relationship with photo synthetic and ammonium assimilating efficiency. Mol Biol Rep 36:2209–2220
Kurai T, Wakayama M, Abiko T, Yanagisawa S, Aoki N, Ohsugi R (2011) Introduction of the ZmDof1 gene into rice enhances carbon and nitrogen assimilation under low-nitrogen conditions. Plant Biotechnol J 9:826–837
Kushwaha H, Gupta S, Singh VK, Rastogi S, Yadav D (2011) Genome wide identification of Dof transcription factor gene family in sorghum and its comparative phylogenetic analysis with rice and Arabidopsis. Mol Biol Rep 38:5037–5053
Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R et al (2007) Clustal W and Clustal X version 2.0. Bioinformatics. Bioinformatics 21:2947–2948
Latchman DS (1997) Transcription factors: an overview. Int J Biochem Cell Biol 29:1305–1312
Li D, Yang C, Li X, Gan Q, Zhao X, Zhu L (2009) Functional characterization of rice OsDof12. Planta 229:1159–1169
Li CH, Cai B, Lou XM, Tang R (2013) Genome-wide analysis of the Dof transcription factor gene family in grape. J Yangzhou Univ (Agric Life Sci Ed) 4:99–103
Lijavetzky D, Carbonero P, Vicente-Carbajosa J (2003) Genome-wide comparative phylogenetic analysis of the rice and Arabidopsis Dof gene families. BMC Evol Biol 3:17
Lin XC, Chow TY, Chen HH, Liu CC, Chou SJ, Huang BL, Kuo CI, Wen CK, Huang LC, Fang W (2010) Understanding bamboo flowering based on large-scale analysis of expressed sequence tags. Genet Mol Res 9:1085–1093
Marzabal P, Gas E, Fontanet P, Vicente-Carbajosa J, Torrent M, Ludevid MD (2008) The maize Dof protein PBF activates transcription of γ-zein during maize seed development. Plant Mol Biol 67:441–454
Mena M, Vicente-Carbajosa J, Schmidt RJ, Carbonero P (1998) An endosperm-specific DOF protein from barley, highly conserved in wheat, binds to and activates transcription from the prolamin-box of a native B-hordein promoter in barley endosperm. Plant J 1:53–62
Mena M, Cejudo FJ, Isabel-Lamoneda I, Carbonero P (2002) A role for the DOF transcription factor BPBF in the regulation of gibberellin-responsive genes in barley aleurone. Plant Physiol 130:111–119
Moreno-Risueno MA, Dıaz I, Carrillo L, Fuentes R, Carbonero P (2007a) The HvDOF19 transcription factor mediates the abscisic acid-dependent repression of hydrolase genes in germinating barley aleurone. Plant J 51:352–365
Moreno-Risueno MÁ, Martínez M, Vicente-Carbajosa J, Carbonero P (2007b) The family of DOF transcription factors: from green unicellular algae to vascular plants. Mol Genet Genom 277:379–390
Papi M, Sabatini S, Bouchez D, Camilleri C, Costantino P, Vittorioso P (2000) Identification and disruption of an Arabidopsis zinc finger gene controlling seed germination. Gene Dev 14:28–33
Park DH, Lim PO, Kim JS, Cho DS, Hong SH, Nam HG (2003) The Arabidopsis COG1 gene encodes a Dof domain transcription factor and negatively regulates phytochrome signaling. Plant J 34:161–171
Peng ZH, Lu TT, Li LB, Liu X, Gao ZM, Hu T, Yang X, Feng Q, Guan J, Weng QJ et al (2010) Genome-wide characterization of the biggest grass, bamboo, based on 10,608 putative full-length cDNA sequences. BMC Plant Biol 10:116
Peng Z, Lu Y, Li L, Zhao Q, Feng Q, Gao ZM, Lu HY, Hu T, Yao N, Liu KY et al (2013) The draft genome of the fast-growing non-timber forest species moso bamboo (Phyllostachys heterocycla). Nature Genet 4:456–463
Plesch G, Ehrhardt T, Mueller-Roeber B (2001) Involvement of TAAAG elements suggests a role for Dof transcription factors in guard cell-specific gene expression. Plant J 4:455–464
Riechmann JL, Heard J, Martin G, Reuber L, Jiang CZ, Keddie J, Adam L, Pineda Ratcliffe J, Samaha RR et al (2000) Arabidopsis transcription factors genome-wide comparative analysis among eukaryotes. Science 290:2105–2110
Rizza A, Boccaccini A, Lopez-Vidriero I, Costantino P, Vittorioso P (2011) Inactivation of the ELIP1 and ELIP2 genes affects Arabidopsis seed germination. New Phytol 190:896–905
Rueda-Lopez M, Crespillo R, Canovas FM, Avila C (2008) Differential regulation of two glutamine synthetase genes by a single Dof transcription factor. Plant J 56:73–85
Rueda-Romero P, Barrero-Sicilia C, Gomez-Cadenas A, Carbonero P, Onate-Sanchez L (2012) Arabidopsis thaliana DOF6 negatively affects germination in non-after-ripened seeds and interacts with TCP14. J Exp Bot 5:1937–1949
Sawa M, Nusinow DA, Kay SA, Imaizumi T (2007) FKF1 and GIGANTEA complex formation is required for day-length measurement in Arabidopsis. Science 318:261–265
Schmutz J, Cannon SB, Schlueter J, Ma J, Mitros T, Nelson W (2010) Genome sequence of the palaeopolyploid soybean. Nature 463:178–183
Shaw LM, McIntyre CL, Gresshoff PM, Xue G (2009) Members of the Dof transcription factor family in Triticum aestivum are associated with light-mediated gene regulation. Funct Integr Genom 9:485–498
Shigyo M, Tabei N, Yoneyama T, Yanagisawa S (2007) Evolutionary processes during the formation of the plant-specific Dof transcription factor family. Plant Cell Physiol 1:179–185
Skirycz A, Reichelt M, Burow M, Birkemeyer C, Rolcik J, Kopka J, Zanor MI, Gershenzon J, Strnad M, Szopa J et al (2006) DOF transcription factor AtDof1.1 (OBP2) is part of a regulatory network controlling glucosinolate biosynthesis in Arabidopsis. Plant J 47:10–24
Skirycz A, Jozefczuk S, Stobiecki M, Muth D, Zanor MI, Witt I, Mueller-Roeber B (2007) Transcription factor AtDOF4.2 affects phenylpropanoid metabolism in Arabidopsis thaliana. New Phytol 175:425–438
Song YH, Smith RW, To BJ, Millar AJ, Imaizumi T (2012) FKF1 conveys crucial timing information for CONSTANS stabilization in the photoperiodic flowering. Science 6084:1045–1049
Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Abiko T, Kumar S (2011) MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony. Mol Biol Evol 10:2731–2739
Tanaka M, Takahata Y, Nakayama H, Nakatan M, Tahara M (2009) Altered carbohydrate metabolism in the storage roots of sweetpotato plants overexpressing the SRF1 gene, which encodes a Dof zinc finger transcription factor. Planta 230:737–746
Vicente-Carbajosa J, Moose SP, Parsons RL, Schmidt RJ (1997) A maize zinc-finger protein binds the prolamin box in zein gene promoters and interacts with the basic leucine zipper transcriptional activator Opaque2. Proc Natl Acad Sci USA 94:7685–7690
Wang H, Zhang B, Hao Y, Huang J, Tian A, Liao Y, Zhang J, Chen S (2007) The soybean Dof-type transcription factor genes, GmDof4 and GmDof11, enhance lipid content in the seeds of transgenic Arabidopsis plants. Plant J 52:716–729
Ward JM, Cufr CA, Denzel MA, Neff MM (2005) The Dof transcription factor OBP3 modulates phytochrome and cryptochrome signaling in Arabidopsis. Plant Cell 17:475–485
Washio K (2003) Functional dissections between GAMYB and Dof transcription factors suggest a role for protein-protein associations in the gibberellin-mediated expression of the RAmy1A gene in the rice aleurone. Plant Physiol 133:850–863
Wei P, Tan F, Gao X, Zhang X, Wang G, Xu H, Li L, Chen J, Wang X (2010) Over-expression of AtDOF4.7, an Arabidopsis DOF family transcription factor, induces floral organ abscission deficiency in Arabidopsis. Plant Physiol 153:1031–1045
Yanagisawa S (1995) A novel DNA-binding domain that may form a single zinc finger motif. Nucleic Acids Res 17:3403–3410
Yanagisawa S (2000) Dof1 and Dof2 transcription factors are associated with expression of multiple genes involved in carbon metabolism in maize. Plant J 3:281–288
Yanagisawa S, Sheen J (1998) Involvement of maize Dof zinc finger proteins in tissue-specific and light-regulated gene expression. Plant Cell 10:75–89
Yang XH, Tuskan GA, Cheng ZM (2006a) Divergence of the dof gene families in poplar, Arabidopsis and rice suggests multiple modes of gene evolution after duplication. Plant Physiol 142:820–830
Yang X, Tuskan GA, Cheng Z (2006b) Divergence of the Dof gene families in poplar, Arabidopsis, and rice suggests multiple modes of gene evolution after duplication. Plant Physiol 142:820–830
Yang J, Yang M, Wang D, Chen F, Shen S (2010) JcDof1 a Dof transcription factor gene, is associated with the light-mediated circadian clock in Jatropha curcas. Physiol Plantarum 139:324–334
Yang J, Yang M, Zhang W, Chen F, Shen S (2011) A putative flowering-time-related Dof transcription factor gene, JcDof3, is controlled by the circadian clock in Jatropha curcas. Plant Sci 181:667–674
Zhang B, Chen W, Foley RC, Büttner M, Singh KB (1995) Interactions between distinct types of DNA binding proteins enhance binding to ocs element promoter. Plant Cell 7:2241–2252
Acknowledgments
This work was supported by the Forestry Project of the Ministry of Science and Technology of the People’s Republic of China (Grant 200704001 to Z. J.), and the Chinese Academy of Sciences (KSCX2-YW-G-034 to B. H.).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
We declare that no competing interests exist.
Additional information
Tao Wang and Jin-Jun Yue have contributed equally to this paper.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Wang, T., Yue, JJ., Wang, XJ. et al. Genome-wide identification and characterization of the Dof gene family in moso bamboo (Phyllostachys heterocycla var. pubescens). Genes Genom 38, 733–745 (2016). https://doi.org/10.1007/s13258-016-0418-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13258-016-0418-2