Abstract
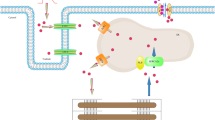
We review the recent development of novel biochemical and spectroscopic methods to determine the site-specific phosphorylation, expression, mutation, and structural dynamics of phospholamban (PLB), in relation to its function (inhibition of the cardiac calcium pump, SERCA2a), with specific focus on cardiac physiology, pathology, and therapy. In the cardiomyocyte, SERCA2a actively transports Ca2+ into the sarcoplasmic reticulum (SR) during relaxation (diastole) to create the concentration gradient that drives the passive efflux of Ca2+ required for cardiac contraction (systole). Unphosphorylated PLB (U-PLB) inhibits SERCA2a, but phosphorylation at S16 and/or T17 (producing P-PLB) changes the structure of PLB to relieve SERCA2a inhibition. Because insufficient SERCA2a activity is a hallmark of heart failure, SERCA2a activation, by gene therapy (Andino et al. 2008; Fish et al. 2013; Hoshijima et al. 2002; Jessup et al. 2011) or drug therapy (Ferrandi et al. 2013; Huang 2013; Khan et al. 2009; Rocchetti et al. 2008; Zhang et al. 2012), is a widely sought goal for treatment of heart failure. This review describes rational approaches to this goal. Novel biophysical assays, using site-directed labeling and high-resolution spectroscopy, have been developed to resolve the structural states of SERCA2a-PLB complexes in vitro and in living cells. Novel biochemical assays, using synthetic standards and multidimensional immunofluorescence, have been developed to quantitate PLB expression and phosphorylation states in cells and human tissues. The biochemical and biophysical properties of U-PLB, P-PLB, and mutant PLB will ultimately resolve the mechanisms of loss of inhibition and gain of inhibition to guide therapeutic development. These assays will be powerful tools for investigating human tissue samples from the Sydney Heart Bank, for the purpose of analyzing and diagnosing specific disorders.










Similar content being viewed by others
References
Ablorh NA, Miller T, Nitu F, Gruber SJ, Karim C, Thomas DD (2012) Accurate quantitation of phospholamban phosphorylation. Immunoblot Anal Biochem 425:68–75. doi:10.1016/j.ab.2012.01.028
Ablorh NA, Dong X, James ZM, Xiong Q, Zhang J, Thomas DD, Karim CB (2014) Synthetic Phosphopeptides Enable Quantitation of the Content and Function of Phospholamban's four Phosphorylation States. Cardiac Muscle J Biol Chem. doi:10.1074/jbc.M114.556621
Abu-Baker S, Lorigan GA (2006) Phospholamban and its phosphorylated form interact differently with lipid bilayers: A 31P, 2H, and 13C solid-state NMR spectroscopic study. Biochemistry 45:13312–13322. doi:10.1021/bi0614028
Afara MR, Trieber CA, Glaves JP, Young HS (2006) Rational design of peptide inhibitors of the sarcoplasmic reticulum calcium pump. Biochemistry 45:8617–8627. doi:10.1021/bi0523761
Andino LM, Takeda M, Kasahara H, Jakymiw A, Byrne BJ, Lewin AS (2008) AAV-mediated knockdown of phospholamban leads to improved contractility and calcium handling in cardiomyocytes. J Gen Med 10:132–142. doi:10.1002/jgm.1131
Asahi M, Kimura Y, Kurzydlowski K, Tada M, MacLennan DH (1999) Transmembrane helix M6 in sarco(endo)plasmic reticulum Ca(2+)-ATPase forms a functional interaction site with phospholamban. Evidence for physical interactions at other sites. J Biol Chem 274:32855–32862
Asahi M, Green NM, Kurzydlowski K, Tada M, MacLennan DH (2001) Phospholamban domain IB forms an interaction site with the loop between transmembrane helices M6 and M7 of sarco(endo)plasmic reticulum Ca2+ ATPases. Proc Natl Acad Sci U S A 98:10061–10066. doi:10.1073/pnas.181348298
Bers DM (2002) Cardiac excitation-contraction coupling. Nature 415:198–205. doi:10.1038/415198a
Boknik P et al (2001) Enhanced protein phosphorylation in hypertensive hypertrophy. Cardiovasc Res 51:717–728
Brittsan AG et al (2003) Chronic SR Ca2 + -ATPase inhibition causes adaptive changes in cellular Ca2+ transport. Circ Res 92:769–776. doi:10.1161/01.RES.0000066661.49920.59
Calaghan S, Kozera L, White E (2008) Compartmentalisation of cAMP-dependent signalling by caveolae in the adult cardiac myocyte. J Mol Cell Cardiol 45:88–92. doi:10.1016/j.yjmcc.2008.04.004
Cantilina T, Sagara Y, Inesi G, Jones LR (1993) Comparative studies of cardiac and skeletal sarcoplasmic reticulum ATPases. Effect of a phospholamban antibody on enzyme activation by Ca2+. J Biol Chem 268:17018–17025
Ceholski DK, Trieber CA, Holmes CF, Young HS (2012) Lethal, hereditary mutants of phospholamban elude phosphorylation by protein kinase A. J Biol Chem 287:26596–26605. doi:10.1074/jbc.M112.382713
Chen Z (2014) Competitive displacement of wild-type phospholamban from the Ca-free cardiac calcium pump by phospholamban mutants with different binding affinities. J Mol Cell Cardiol 76C:130–137. doi:10.1016/j.yjmcc.2014.08.020
Chen Z, Stokes DL, Jones LR (2005) Role of leucine 31 of phospholamban in structural and functional interactions with the Ca2+ pump of cardiac sarcoplasmic reticulum. J Biol Chem 280:10530–10539. doi:10.1074/jbc.M414007200
Chu G, Lester JW, Young KB, Luo W, Zhai J, Kranias EG (2000) A single site (Ser16) phosphorylation in phospholamban is sufficient in mediating its maximal cardiac responses to beta -agonists. J Biol Chem 275:38938–38943. doi:10.1074/jbc.M004079200
Cornea RL, Jones LR, Autry JM, Thomas DD (1997) Mutation and phosphorylation change the oligomeric structure of phospholamban in lipid bilayers. Biochemistry 36:2960–2967. doi:10.1021/bi961955q
Cornea RL et al (2013) High-throughput FRET assay yields allosteric SERCA2a activators. J Biomol Screen 18:97–107. doi:10.1177/1087057112456878
De Simone A, Gustavsson M, Montalvao RW, Shi L, Veglia G, Vendruscolo M (2013) Structures of the excited states of phospholamban and shifts in their populations upon phosphorylation. Biochemistry 52:6684–6694. doi:10.1021/bi400517b
Dong X, Thomas DD (2014) Time-resolved FRET reveals the structural mechanism of SERCA2a-PLB regulation. Biochem Biophys Res Commun 449:196–201. doi:10.1016/j.bbrc.2014.04.166
Fechner H et al (2007) Highly efficient and specific modulation of cardiac calcium homeostasis by adenovector-derived short hairpin RNA targeting phospholamban. Gene Ther 14:211–218. doi:10.1038/sj.gt.3302872
Ferrandi M et al (2013) Istaroxime stimulates SERCA2a and accelerates calcium cycling in heart failure by relieving phospholamban inhibition. Br J Pharmacol 169:1849–1861. doi:10.1111/bph.12278
Fish KM et al (2013) AAV9.I-1c delivered via direct coronary infusion in a porcine model of heart failure improves contractility and mitigates adverse remodeling. Circ Heart Fail 6:310–317. doi:10.1161/CIRCHEARTFAILURE.112.971325
Grote-Wessels S et al (2008) Inhibition of protein phosphatase 1 by inhibitor-2 exacerbates progression of cardiac failure in a model with pressure overload. Cardiovasc Res 79:464–471. doi:10.1093/cvr/cvn113
Gruber SJ, Haydon S, Thomas DD (2012) Phospholamban mutants compete with wild type for SERCA2a binding in living cells. Biochem Biophys Res Commun 420:236–240. doi:10.1016/j.bbrc.2012.02.125
Gruber SJ et al (2014) Discovery of enzyme modulators via high-throughput time-resolved FRET in living cells. J Biomol Screen 19:215–222. doi:10.1177/1087057113510740
Guinto PJ, Haim TE, Dowell-Martino CC, Sibinga N, Tardiff JC (2009) Temporal and mutation-specific alterations in Ca2+ homeostasis differentially determine the progression of cTnT-related cardiomyopathies in murine models. Am J Physiol Heart Circ Physiol 297:H614–626. doi:10.1152/ajpheart.01143.2008
Gustavsson M, Traaseth NJ, Karim CB, Lockamy EL, Thomas DD, Veglia G (2011) Lipid-mediated folding/unfolding of phospholamban as a regulatory mechanism for the sarcoplasmic reticulum Ca2 + -ATPase. J Mol Biol 408:755–765. doi:10.1016/j.jmb.2011.03.015
Gustavsson M, Verardi R, Mullen DG, Mote KR, Traaseth NJ, Gopinath T, Veglia G (2013) Allosteric regulation of SERCA2a by phosphorylation-mediated conformational shift of phospholamban. Proc Natl Acad Sci U S A 110:17338–17343. doi:10.1073/pnas.1303006110
Gwathmey JK et al (1991) Diastolic dysfunction in hypertrophic cardiomyopathy. Effect on active force generation during systole. J Clin Invest 87:1023–1031. doi:10.1172/JCI115061
Ha KN et al (2007) Controlling the inhibition of the sarcoplasmic Ca2 + -ATPase by tuning phospholamban structural dynamics. J Biol Chem 282:37205–37214. doi:10.1074/jbc.M704056200
Ha KN, Gustavsson M, Veglia G (2012) Tuning the structural coupling between the transmembrane and cytoplasmic domains of phospholamban to control sarcoplasmic reticulum Ca(2+)-ATPase (SERCA2a) function. J Muscle Res Cell Motil 33:485–492. doi:10.1007/s10974-012-9319-4
Haghighi K et al (2003) Human phospholamban null results in lethal dilated cardiomyopathy revealing a critical difference between mouse and human. J Clin Invest 111:869–876. doi:10.1172/JCI17892
Haghighi K et al (2012) The human phospholamban Arg14-deletion mutant localizes to plasma membrane and interacts with the Na/K-ATPase. J Mol Cell Cardiol 52:773–782. doi:10.1016/j.yjmcc.2011.11.012
He H et al (1999) Effects of mutant and antisense RNA of phospholamban on SR Ca(2+)-ATPase activity and cardiac myocyte contractility. Circulation 100:974–980
Hoshijima M et al (2002) Chronic suppression of heart-failure progression by a pseudophosphorylated mutant of phospholamban via in vivo cardiac rAAV gene delivery. Nat Med 8:864–871. doi:10.1038/nm739
Hou Z, Kelly EM, Robia SL (2008) Phosphomimetic mutations increase phospholamban oligomerization and alter the structure of its regulatory complex. J Biol Chem 283:28996–29003. doi:10.1074/jbc.M804782200
Huang CL (2013) SERCA2a stimulation by istaroxime: a novel mechanism of action with translational implications. Br J Pharmacol 170:486–488. doi:10.1111/bph.12288
Huke S, Periasamy M (2004) Phosphorylation-status of phospholamban and calsequestrin modifies their affinity towards commonly used antibodies. J Mol Cell Cardiol 37:795–799. doi:10.1016/j.yjmcc.2004.06.003
Iwanaga Y et al (2004) Chronic phospholamban inhibition prevents progressive cardiac dysfunction and pathological remodeling after infarction in rats. J Clin Invest 113:727–736. doi:10.1172/JCI18716
James ZM, McCaffrey JE, Torgersen KD, Karim CB, Thomas DD (2012) Protein-protein interactions in calcium transport regulation probed by saturation transfer electron paramagnetic resonance. Biophys J 103:1370–1378. doi:10.1016/j.bpj.2012.08.032
Jessup M et al (2011) Calcium Upregulation by Percutaneous Administration of Gene Therapy in Cardiac Disease (CUPID): a phase 2 trial of intracoronary gene therapy of sarcoplasmic reticulum Ca2 + -ATPase in patients with advanced heart failure. Circulation 124:304–313. doi:10.1161/CIRCULATIONAHA.111.022889
Ji Y, Loukianov E, Loukianova T, Jones LR, Periasamy M (1999) SERCA2a1a can functionally substitute for SERCA2a in the heart. Am J Physiol 276:H89–97
Karim CB, Stamm JD, Karim J, Jones LR, Thomas DD (1998) Cysteine reactivity and oligomeric structures of phospholamban and its mutants. Biochemistry 37:12074–12081. doi:10.1021/bi980642n
Karim CB, Zhang Z, Howard EC, Torgersen KD, Thomas DD (2006) Phosphorylation-dependent conformational switch in spin-labeled phospholamban bound to SERCA2a. J Mol Biol 358:1032–1040. doi:10.1016/j.jmb.2006.02.051
Kaye DM et al (2007) Percutaneous cardiac recirculation-mediated gene transfer of an inhibitory phospholamban peptide reverses advanced heart failure in large animals. J Am Coll Cardiol 50:253–260. doi:10.1016/j.jacc.2007.03.047
Kelly EM, Hou Z, Bossuyt J, Bers DM, Robia SL (2008) Phospholamban oligomerization, quaternary structure, and sarco(endo)plasmic reticulum calcium ATPase binding measured by fluorescence resonance energy transfer in living cells. J Biol Chem 283:12202–12211. doi:10.1074/jbc.M707590200
Khan H et al (2009) Istaroxime, a first in class new chemical entity exhibiting SERCA2a-2 activation and Na-K-ATPase inhibition: a new promising treatment for acute heart failure syndromes. Heart Fail Rev 14:277–287. doi:10.1007/s10741-009-9136-z
Kimura Y, Kurzydlowski K, Tada M, MacLennan DH (1997) Phospholamban inhibitory function is activated by depolymerization. J Biol Chem 272:15061–15064
Kranias EG, Hajjar RJ (2012) Modulation of cardiac contractility by the phospholamban/SERCA2a regulatome. Circ Res 110:1646–1660. doi:10.1161/CIRCRESAHA.111.259754
Li J, Boschek CB, Xiong Y, Sacksteder CA, Squier TC, Bigelow DJ (2005) Essential role for Pro21 in phospholamban for optimal inhibition of the Ca-ATPase. Biochemistry 44:16181–16191. doi:10.1021/bi051075o
Li J, James ZM, Dong X, Karim CB, Thomas DD (2012) Structural and functional dynamics of an integral membrane protein complex modulated by lipid headgroup charge. J Mol Biol 418:379–389. doi:10.1016/j.jmb.2012.02.011
Li A et al (2013) Heart research advances using database search engines. Human Protein Atlas and the Sydney Heart Bank. Heart Lung Circ 22:819–826. doi:10.1016/j.hlc.2013.06.006
Lockamy EL, Cornea RL, Karim CB, Thomas DD (2011) Functional and physical competition between phospholamban and its mutants provides insight into the molecular mechanism of gene therapy for heart failure. Biochem Biophys Res Commun 408:388–392. doi:10.1016/j.bbrc.2011.04.023
Loyer X et al (2008) Cardiomyocyte overexpression of neuronal nitric oxide synthase delays transition toward heart failure in response to pressure overload by preserving calcium cycling. Circulation 117:3187–3198. doi:10.1161/CIRCULATIONAHA.107.741702
MacLennan DH, Toyofuku T, Kimura Y (1997) Sites of regulatory interaction between calcium ATPases and phospholamban. Basic Res Cardiol 92(Suppl 1):11–15
Mandinov L, Eberli FR, Seiler C, Hess OM (2000) Diastolic heart failure. Cardiovasc Res 45:813–825
Marks AR (2013) Calcium cycling proteins and heart failure: Mechanisms and therapeutics. J Clin Invest 123:46–52. doi:10.1172/JCI62834
Masterson LR, Yu T, Shi L, Wang Y, Gustavsson M, Mueller MM, Veglia G (2011) cAMP-dependent protein kinase A selects the excited state of the membrane substrate phospholamban. J Mol Biol 412:155–164. doi:10.1016/j.jmb.2011.06.041
Mayer EJ, Huckle W, Johnson RG Jr, McKenna E (2000) Characterization and quantitation of phospholamban and its phosphorylation state using antibodies. Biochem Biophys Res Commun 267:40–48. doi:10.1006/bbrc.1999.1920
Medeiros A et al (2011) Mutations in the human phospholamban gene in patients with heart failure. Am Heart J 162(1088–1095):e1081. doi:10.1016/j.ahj.2011.07.028
Metcalfe EE, Zamoon J, Thomas DD, Veglia G (2004) (1)H/(15)N heteronuclear NMR spectroscopy shows four dynamic domains for phospholamban reconstituted in dodecylphosphocholine micelles. Biophys J 87:1205–1214. doi:10.1529/biophysj.103.038844
Metcalfe EE, Traaseth NJ, Veglia G (2005) Serine 16 phosphorylation induces an order-to-disorder transition in monomeric phospholamban. Biochemistry 44:4386–4396. doi:10.1021/bi047571e
Mills GD, Kubo H, Harris DM, Berretta RM, Piacentino V 3rd, Houser SR (2006) Phosphorylation of phospholamban at threonine-17 reduces cardiac adrenergic contractile responsiveness in chronic pressure overload-induced hypertrophy. Am J Physiol Heart Circ Physiol 291:H61–70. doi:10.1152/ajpheart.01353.2005
Miyazaki Y et al (2012) Heart failure-inducible gene therapy targeting protein phosphatase 1 prevents progressive left ventricular remodeling. PLoS ONE 7:e35875. doi:10.1371/journal.pone.0035875
Mueller B, Karim CB, Negrashov IV, Kutchai H, Thomas DD (2004) Direct detection of phospholamban and sarcoplasmic reticulum Ca-ATPase interaction in membranes using fluorescence resonance energy transfer. Biochemistry 43:8754–8765. doi:10.1021/bi049732k
Nesmelov YE, Karim CB, Song L, Fajer PG, Thomas DD (2007) Rotational dynamics of phospholamban determined by multifrequency electron paramagnetic resonance. Biophys J 93:2805–2812. doi:10.1529/biophysj.107.108910
Oh JG et al (2013) Decoy peptides targeted to protein phosphatase 1 inhibit dephosphorylation of phospholamban in cardiomyocytes. J Mol Cell Cardiol 56:63–71. doi:10.1016/j.yjmcc.2012.12.005
Pritchard TJ et al (2013) Active inhibitor-1 maintains protein hyper-phosphorylation in aging hearts and halts remodeling in failing hearts. PLoS ONE 8:e80717. doi:10.1371/journal.pone.0080717
Reue K (1998) mRNA quantitation techniques: Considerations for experimental design and application. J Nutr 128:2038–2044
Rocchetti M et al (2008) Modulation of sarcoplasmic reticulum function by PST2744 [istaroxime; (E, Z)-3-((2-aminoethoxy)imino) androstane-6,17-dione hydrochloride)] in a pressure-overload heart failure model. J Pharmacol Exp Ther 326:957–965. doi:10.1124/jpet.108.138701
Santana LF, Kranias EG, Lederer WJ (1997) Calcium sparks and excitation-contraction coupling in phospholamban-deficient mouse ventricular myocytes. J Physiol 503(Pt 1):21–29
Schmidt AG, Edes I, Kranias EG (2001) Phospholamban: A promising therapeutic target in heart failure. Cardiovasc Drugs Ther 15:387–396
Schmitt JP et al (2003) Dilated cardiomyopathy and heart failure caused by a mutation in phospholamban. Science 299:1410–1413. doi:10.1126/science.1081578
Simmerman HK, Kobayashi YM, Autry JM, Jones LR (1996) A leucine zipper stabilizes the pentameric membrane domain of phospholamban and forms a coiled-coil pore structure. J Biol Chem 271:5941–5946
Smith CJ, Osborn AM (2009) Advantages and limitations of quantitative PCR (Q-PCR)-based approaches in microbial ecology. FEMS Microbiol Ecol 67:6–20. doi:10.1111/j.1574-6941.2008.00629.x
Steenaart NA, Ganim JR, Di Salvo J, Kranias EG (1992) The phospholamban phosphatase associated with cardiac sarcoplasmic reticulum is a type 1 enzyme. Arch Biochem Biophys 293:17–24
Toyofuku T, Kurzydlowski K, Tada M, MacLennan DH (1994) Amino acids Glu2 to Ile18 in the cytoplasmic domain of phospholamban are essential for functional association with the Ca(2+)-ATPase of sarcoplasmic reticulum. J Biol Chem 269:3088–3094
Traaseth NJ, Verardi R, Torgersen KD, Karim CB, Thomas DD, Veglia G (2007) Spectroscopic validation of the pentameric structure of phospholamban. Proc Natl Acad Sci U S A 104:14676–14681. doi:10.1073/pnas.0701016104
Traaseth NJ, Shi L, Verardi R, Mullen DG, Barany G, Veglia G (2009) Structure and topology of monomeric phospholamban in lipid membranes determined by a hybrid solution and solid-state NMR approach. Proc Natl Acad Sci U S A 106:10165–10170. doi:10.1073/pnas.0904290106
Trieber CA, Douglas JL, Afara M, Young HS (2005) The effects of mutation on the regulatory properties of phospholamban in co-reconstituted membranes. Biochemistry 44:3289–3297. doi:10.1021/bi047878d
Trieber CA, Afara M, Young HS (2009) Effects of phospholamban transmembrane mutants on the calcium affinity, maximal activity, and cooperativity of the sarcoplasmic reticulum calcium pump. Biochemistry 48:9287–9296. doi:10.1021/bi900852m
Tsuji T et al (2009) Rescue of Ca2+ overload-induced left ventricular dysfunction by targeted ablation of phospholamban. Am J Physiol Heart Circ Physiol 296:H310–317. doi:10.1152/ajpheart.00975.2008
Vostrikov VV, Mote KR, Verardi R, Veglia G (2013) Structural dynamics and topology of phosphorylated phospholamban homopentamer reveal its role in the regulation of calcium transport. Structure 21:2119–2130. doi:10.1016/j.str.2013.09.008
Wang Y, Goldhaber JI (2004) Return of calcium: Manipulating intracellular calcium to prevent cardiac pathologies. Proc Natl Acad Sci U S A 101:5697–5698. doi:10.1073/pnas.0401518101
Wegener AD, Simmerman HK, Lindemann JP, Jones LR (1989) Phospholamban phosphorylation in intact ventricles. Phosphorylation of serine 16 and threonine 17 in response to beta-adrenergic stimulation. J Biol Chem 264:11468–11474
Xu XL, Ji H, Gu SY, Shao Q, Huang QJ, Cheng YP (2007) Modification of alterations in cardiac function and sarcoplasmic reticulum by astragaloside IV in myocardial injury in vivo. Eur J Pharmacol 568:203–212. doi:10.1016/j.ejphar.2007.04.007
Yu X, Lorigan GA (2014) Secondary structure, backbone dynamics, and structural topology of phospholamban and its phosphorylated and Arg9Cys-mutated forms in phospholipid bilayers utilizing 13C and 15 N solid-state NMR spectroscopy. J Phys Chem B 118:2124–2133. doi:10.1021/jp500316s
Zamoon J, Mascioni A, Thomas DD, Veglia G (2003) NMR solution structure and topological orientation of monomeric phospholamban in dodecylphosphocholine micelles. Biophys J 85:2589–2598. doi:10.1016/S0006-3495(03)74681-5
Zhang Z, Lewis D, Strock C, Inesi G, Nakasako M, Nomura H, Toyoshima C (2000) Detailed characterization of the cooperative mechanism of Ca(2+) binding and catalytic activation in the Ca(2+) transport (SERCA2a) ATPase. Biochemistry 39:8758–8767
Zhang DW et al (2012) Astragaloside IV alleviates hypoxia/reoxygenation-induced neonatal rat cardiomyocyte injury via the protein kinase a pathway. Pharmacology 90:95–101. doi:10.1159/000339476
Zhao W et al (2003) Combined phospholamban ablation and SERCA2a1a overexpression result in a new hyperdynamic cardiac state. Cardiovasc Res 57:71–81
Ziolo MT, Martin JL, Bossuyt J, Bers DM, Pogwizd SM (2005) Adenoviral gene transfer of mutant phospholamban rescues contractile dysfunction in failing rabbit myocytes with relatively preserved SERCA2a function. Circ Res 96:815–817. doi:10.1161/01.RES.0000163981.97262.3b
Acknowledgments
This project used the facilities of the Biophysical Spectroscopy Center and the Peptide Synthesis Facility, University of Minnesota. We thank Simon J. Gruber for expert advice and Octavian Cornea for preparing the manuscript. Many others contributed to the studies reviewed here, especially Christine B. Karim, Razvan L. Cornea, Jesse E. McCaffrey, Zachary M. James, and Xiaoqiong Dong.
Compliance with ethical standards
ᅟ
Funding
This work was supported by National Institutes of Health grants to D.D.T. (AG26160, AG042996) and N.D.A. (T32 HL069764).
Conflict of interest
Naa-Adjeley Dromoh Ablorh declares that she has no conflict of interest. David Dale Thomas declares that he has no conflict of interest.
Ethical approval
This article does not contain any studies with animals performed by any of the authors.
Author information
Authors and Affiliations
Corresponding author
Additional information
Special Issue: Biophysics of Human Heart Failure
Rights and permissions
About this article
Cite this article
Ablorh, NA.D., Thomas, D.D. Phospholamban phosphorylation, mutation, and structural dynamics: a biophysical approach to understanding and treating cardiomyopathy. Biophys Rev 7, 63–76 (2015). https://doi.org/10.1007/s12551-014-0157-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12551-014-0157-z