Abstract
Producing cellulosic biofuels from plant material has recently emerged as a key US Department of Energy goal. For this technology to be commercially viable on a large scale, it is critical to make production cost efficient by streamlining both the deconstruction of lignocellulosic biomass and fuel production. Many natural ecosystems efficiently degrade lignocellulosic biomass and harbor enzymes that, when identified, could be used to increase the efficiency of commercial biomass deconstruction. However, ecosystems most likely to yield relevant enzymes, such as tropical rain forest soil in Puerto Rico, are often too complex for enzyme discovery using current metagenomic sequencing technologies. One potential strategy to overcome this problem is to selectively cultivate the microbial communities from these complex ecosystems on biomass under defined conditions, generating less complex biomass-degrading microbial populations. To test this premise, we cultivated microbes from Puerto Rican soil or green waste compost under precisely defined conditions in the presence dried ground switchgrass (Panicum virgatum L.) or lignin, respectively, as the sole carbon source. Phylogenetic profiling of the two feedstock-adapted communities using SSU rRNA gene amplicon pyrosequencing or phylogenetic microarray analysis revealed that the adapted communities were significantly simplified compared to the natural communities from which they were derived. Several members of the lignin-adapted and switchgrass-adapted consortia are related to organisms previously characterized as biomass degraders, while others were from less well-characterized phyla. The decrease in complexity of these communities make them good candidates for metagenomic sequencing and will likely enable the reconstruction of a greater number of full-length genes, leading to the discovery of novel lignocellulose-degrading enzymes adapted to feedstocks and conditions of interest.
Similar content being viewed by others
Introduction
The US Department of Energy has recently made alternative liquid fuel production from lignocellulosic biomass a primary goal. Establishing such renewable, low-carbon liquid fuel alternatives is a critical short- and long-term solution to the environmental problems and national security risks associated with petroleum consumption. Cellulosic biofuels are one such alternative that are receiving unprecedented international attention, owing to the large, underutilized reservoir of renewable energy in plant biomass [10, 30]. Currently, one of the major barriers to the large-scale production of inexpensive cellulosic biofuels is the ability to efficiently deconstruct biomass into fermentable carbon sources, such as glucose and xylose. Enzymatic saccharification of the plant cell polymers cellulose and hemicellulose is an efficient method to obtain these sugars from biomass, but this process is costly using present-day fungal commercial enzyme cocktails. Discovery of more efficient and robust biomass-degrading enzymes will drive down costs and increase the economic viability of this technology.
Many natural ecosystems, such as soils and compost, almost completely mineralize plant biomass. The indigenous microbes in these ecosystems may provide a rich reservoir of genes relevant to the development of cellulosic biofuels. Target genes include glycosyl hydrolases, enzymes that convert simple sugar intermediates into biofuels [30], and lignolytic enzymes that can either release cellulose from the plant polymer lignin to increase sugar yields from biomass, or facilitate lignin transformation to biobased products. Lignin is of special interest, since currently it is a waste stream in cellulosic biofuels production that is burned to recover heat [24]. Our research, within the Microbial Communities Group, Deconstruction Division, US DOE Joint BioEnergy Institute (JBEI), focuses on two natural biomass-degrading ecosystems: the tropical forest soils of Puerto Rico and municipal green waste compost. Wet tropical forest soils are some of the most productive and diverse terrestrial ecosystems on earth. A recent study identified tropical forest soils as the fastest decomposing soils of plant material compared to all other biomes globally [39]. Green waste compost is another ecosystem where microorganisms rapidly break down lignocellulosic biomass into carbon dioxide, water, and humus. This degradation is so fast, in fact, that the compost heap can heat to 60–70°C, due to the metabolic activity of the microbial community. We are using metagenomics, proteomics, and transcriptomics to investigate these communities, both in their native state and after cultivation on candidate bioenergy feedstocks (Fig. 1).
Schematic structure of our approach to metabolic analysis of complex microbial communities. Starting with natural samples at the bottom, a series of analytical, biochemical, and bioinformatics tools are applied so that eventually we are left with the most active, relevant consortia for novel enzyme and pathway discovery
Identifying specific genes from these ecosystems, which have a high degree of microbial diversity, is challenging. Fortunately, next-generation sequencing technologies such as 454 pyrosequencing can facilitate the discovery of relevant genes [30]. Recent metagenome studies have demonstrated that it is possible to assign functional annotations to partial gene sequences from shotgun sequence reads with a reasonable degree of accuracy, based on BLASTX hits against reference databases [38, 45]. Such annotation can provide a useful functional profile of a community and help identify gene categories of interest. However, this study and others [1] indicate that shotgun metagenome sequence data from highly complex natural microbial communities is of limited use for targeted enzyme discovery, because of the lack of contiguous sequences (contigs) large enough to contain complete open reading frames (ORFs); for cellulases, this is at least 1 kb [33]. For example, Allgaier et al. [1] found only 25 potentially full-length lignocellulose-degrading enzymes from a switchgrass compost microbial community. To discover and extract a greater number of novel functional enzymes for bioenergy applications, lower complexity metagenomic data sets that are more amenable to assembly are required. One possible method for generating less complex microbial communities is to adapt environmental communities to feedstocks (e.g., switchgrass, or lignin) under fixed conditions, such as temperature, pH, etc. that are more directly relevant to the feedstocks and process conditions expected for large-scale industrial biomass deconstruction, with the expectation that the resulting communities will be enriched for microbes expressing desired enzymes.
In this manuscript, we present the results of a shotgun sequencing by 454 Titanium technology of tropical forest soils, which were sufficiently complex to resist assembly of full-length genes. While we present analysis of known lignocellulolytic enzymes and the prospects for enzyme discovery with this data set, the lack of assembly demonstrates the need for less complex communities. To test the premise that feedstock-adapted communities reduce diversity while preserving function, Puerto Rican tropical forest soil and municipal green waste compost microbial communities were adapted to switchgrass anaerobically and lignin aerobically, respectively. Small subunit (SSU) rRNA profiling of both communities demonstrated that they were less complex than the starting inocula and enriched for organisms that we predict are more suited to degrading the target feedstock.
Materials and Methods
Environmental Samples
Tropical forest soil samples were collected in a subtropical lower montane wet forest in the Luquillo Experimental Forest, which is part of the NSF-sponsored Long-Term Ecological Research program in Puerto Rico (18°18′N, 65°50′W). The dominant plant cover is Dacryodes excelsa, with plant species richness of 50 ha−1 made up mostly of early sucessional trees, herbs, and tree ferns. Climate is relatively aseasonal, with mean annual rainfall of 4500 mm and mean annual temperatures of 22 to 24°C [36, 49]. Soils are acidic (pH 5.5), clayey ultisols with high iron and aluminum content and characterized by redox fluctuating from oxic to anoxic on a scale of weeks [36, 42]. Soils were collected from the Bisley watershed ∼250 meters above sea level (masl) from the 0–10 cm depth, using a 2.5-cm diameter soil corer. Cores were stored intact in Ziploc bags at ambient temperature and immediately transported to the lab, where they were used for growth inocula or frozen in liquid nitrogen and stored at −80°C for molecular analysis. Green waste compost inoculum was obtained from a Grover Soil Solutions compost facility located in Zamora, CA, on August 6, 2007. Details of sampling and storage are described in Allgaier et al [1].
DNA Extraction
DNA from the rain forest soil was used for metagenomic sequencing and SSU rRNA profiling; DNA from the green waste compost and the switchgrass- or lignin-adapted cultures was used only for SSU rRNA profiling. DNA was extracted using a modified CTAB extraction method with bead beating and phenol–chloroform extraction, as previously described [6], with the following exceptions: phosphate buffer concentration was 250 mM; 50 µL 100 mM aluminum ammonium sulfate (Sigma Aldrich, St. Louis, MO) was added to the soils as a flocculant for excess humics [5]; DNA was precipitated in Peg6000 solution (30% (w/v) polyethylene glycol 6000 (Sigma Aldrich, St. Louis, MO) in 1.6 M NaCl); soils were extracted a second time as above. These crude extractions were treated further using the Qiagen DNA RNA AllPrep kit (Qiagen, Valencia, CA).
Metagenome Sequencing and Analysis
The native Puerto Rican rainforest soil microbial community was prepared for metagenomic sequencing. Genomic DNA was extracted from the rain forest soil sample and used for sequencing library construction following the DOE Joint Genome Institute standard operating procedure for shotgun sequencing using the Roche 454 GS FLX Titanium technology (Branford, CT). Obtained sequencing reads were quality trimmed using the software tool “lucy” [11] to an accuracy of 99.3%. The unassembled metagenomic data set was loaded into MG-RAST [37] for functional analysis, and subsystem-based metabolic profiles were computed at a maximum E value of 1e−10. We used the November 2008 release of PRIAM [12], modified for nucleotide queries using the “-p F” flag of rpsblast, to assign four-digit EC (Enzyme Commission) numbers, at E < 1e−10. Finally, we used BLASTX to search for homologs (again, at E < 1e−10) against 87,000 enzyme sequences in a local copy of the CAZy [9] and FOLy [31] databases. We used the best BLASTX hit for each sequence read to assign protein family memberships, and also used the best BLASTX hit against the 6,367 CAZy and FOLy enzymes that have independently validated EC numbers to assign a putative EC number to the sequence read (note that many of the protein families in CAZy are highly multifunctional, so in general it is difficult to assign enzyme function based merely on family membership.)
Cultivation Conditions
For adaptation to growth on feedstocks as sole carbon source, tropical forest soil cores were homogenized and inoculated in basal salts minimal medium (BMM) [43] containing trace minerals [44, 50], vitamins [25], and buffered to pH 5.5 to match the measured soil pH using MES. This medium was used for all enrichments (Table 1). Soils were added at 0.5 g per 200 mL BMM, and the resulting mixture was incubated anaerobically at ambient temperatures for 8 weeks with 10 g L−1 dried, ground switchgrass as the sole carbon source. Samples of switchgrass (MPV 2 cultivar) were kindly provided by the laboratory of Dr. Ken Vogel (USDA, ARS, Lincoln, NE).
For the lignin-adapted compost consortia, 100 mg of compost was added to 50 mL of M9TE medium [34] amended with 0.5 g/L lignin in a 250-mL shaker flask and shaken at 200 rpm for 2 weeks at 37°C. Three types of lignin were used in the enrichment cultures: (1) AL—Alkali lignin with low sulfonate content (Sigma Aldrich cat. No. 471003, St. Louis, MO), (2) OL—Organosolv lignin, propionate (Sigma Aldrich cat. No. 371033, St. Louis, MO), (3) IL—Indulin AT (Meadwestvaco, Richmond, VA) that had been washed with water to remove soluble lignin by Soxhlet extraction, following the protocol outlined in Giroux et al. [21]. The water-soluble AL is sulfonated with a molecular weight (MW) range between 10,000 and 60,000; the OL is a mixture of soluble and insoluble lignin; and the IL is unsulfonated, completely insoluble, and likely contains only high MW lignin.
Two milliliters of this culture were then seeded into a fresh flask containing 50 mL of M9TE amended with 0.5 g/L lignin in a 250-mL shaker flask shaken at 200 rpm for 2 weeks at 37°C. This step was repeated three times. After the fifth enrichment culture, cells were prepared for DNA extraction; culture supernatant (10 mL) was placed into five 2-ml microcentrifuge tubes and spun at maximum speed for 5 min. The pellets from each tube were combined and transferred to a 2-mL lysing matrix E tube (MP Biomedicals) and frozen at −80°C.
The M9TE medium was prepared using the following recipe per liter: 200 mL 5× M9 minimal salts, 2 mL 1 M MgCl2, 100 µL 1 M CaCl2, and 60 mL 17× trace elements (final concentration: 470 µM nitrilotriacetic acid, 730 µM MgSO4·7H2O, 180 µM MnSO4·H2O, 1 mM NaCl 1 g, 33 µM FeCl2, 39 µM CoSO4, 41 µM CaCl2·2H2O, 21 µM ZnSO4·7H2O, 2.4 µM CuSO4·5H2O, 1.3 µM AlK(SO4)2·12H2O, 97 µM H3BO3, 2.7 µM Na2MoO4·H2O). The pH was then adjusted 6.5 with KOH. M9TE medium was filter sterilized through a 0.2-µm filter.
SSU ribosomal RNA profiles using PhyloChip Gene Microarray and Amplicon Pyrosequencing
PhyloChip and amplicon pyrosequencing were used to generate SSU rRNA profiles of native and adapted soil communities and native and adapted compost communities, respectively. For PhyloChip analysis, purified genomic DNA was quantified and 30 ng was added to a PCR reaction for amplification of the SSU ribosomal RNA genes. Primers used were 8F for bacteria, 4Fa for archaea, and the same reverse primer for both, 1492R [23, 51]: PCR amplification was performed as previously described [14]. For application onto the high-density SSU rDNA microarray (PhyloChip), PCR products were concentrated to 500 ng (bacteria) or 200 ng (archaea), then pooled, fragmented, biotin-labeled, and hybridized as previously described [6]. For amplicon pyrosequencing, SSU rRNA gene sequences were amplified using the primer pair 926f/1392r, as described in Kunin et al. [28]. The reverse primer included a 5-bp barcode for multiplexing of samples during sequencing. Emulsion PCR and sequencing of the PCR amplicons were performed following manufacturer's instructions for the Roche 454 GS FLX Titanium technology (Branford, CT), with the exception that the final dilution was 1e−8. Sequencing tags were analyzed using the software tool PyroTagger (http://pyrotagger.jgi-psf.org/) using a 395-bp sequence length threshold.
Results
Native Lignocellulolytic Microbial Community Metagenomics
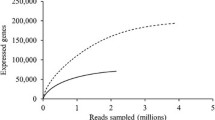
In this study, we used metagenomics to identify enzymes from the native microbial community present in a tropical rain forest soil sample from Puerto Rico (Allgaier et al. analyzed the metagenome of a switchgrass compost in a previously published study [1]). We performed shotgun sequencing using the Roche 454 GS FLX Titanium technology to obtain metagenomic data for the native soil community, resulting in 863,759 reads, with an average read length of 417 bp, for a total of 350Mbp of sequence data. After trimming and quality control, the final data set resulted in 780,588 reads equaling 321 Mbp. Assembly of the tropical forests soil metagenome was attempted using the Newbler assembler software by 454 Life Sciences, but the species composition of the sample was too complex to yield any significant assembly of the metagenome sequence reads. MG-RAST was able to assign various degrees of functional annotation to 29.7% of the sequences (232,025) at E < 1e−10. PRIAM enzyme-specific sequence profiles assigned four-digit EC numbers to 110,411 sequence reads at E < 1e−10. BLASTX of the rainforest metagenome to the CAZy and FOLy databases resulted in 29,051 protein family assignments, and 9,041 EC number assignments (Fig. 2), also at E < 1e−10.
Metabolic profiling of the metagenomic sequence data revealed a wide diversity of protein sequences relevant to bioenergy, including lignocellulolytic enzymes (cellulases, hemicellulases, lignases) as well as potential biofuel synthesis pathways (e.g., mevalonate and non-mevalonate (DOXP) isoprenoid biosynthesis, lycopene biosynthesis, and a proposed butanol biosynthesis pathway). Note that our BLASTX results cover only the lignocellulolytic enzymes present in CAZy and FOLy, whereas PRIAM EC number predictions cover virtually all known metabolic pathways
The most abundant carbohydrate and lignin active enzyme families, as inferred by best BLASTX hits against CAZy and FOLy, are glycoside hydrolases (GH, 12,193 BLASTX hits) and glycosyl transferases (GT, 11,562 hits), followed by carbohydrate esterases (CE, 3,133 hits), lignin oxidases (LO, 413 hits), polysaccharide lyases (PL, 282 hits), and lignin-degrading auxiliary oxidases (LDA, 240 hits; Fig. 3). GT2 and GT4 are large, predominantly bacterial, multifunctional enzyme families, and most of the sequences present here seem to be involved in aspects of bacterial cell-wall biogenesis. GH13 contains mostly starch- and glycogen-degrading enzymes, as well as trehalose synthases. Lignolytic enzymes are relatively low in abundance (likely due to the smaller number of known reference enzymes, and the lack of bacterial sequences in FOLy), consisting mainly of putative cellobiose dehydrogenases (LO3, 361 hits), and a small number of aryl-alcohol oxidases (LDA1, 124 hits), glucose oxidases (LDA6, 82 hits) and laccases (LO1, 47 hits). Not shown are 3,645 hits against enzymes with carbohydrate-binding modules (CBM), the two most abundant of which are CBM13 (previously known as cellulose-binding domain family XIII, 1366 hits) and the glycogen-binding CBM48 (826 hits).
A pie chart demonstrating the relative abundance of CAZy and FOLy enzyme families found in the tropical forest metagenome, inferred from BLASTX hits. GH glycoside hydrolases, GT glycosyl transferrases, PL polysaccharide lyases, CE carbohydrate esterases, LO lignin oxidases, LDA lignin-degrading auxiliary oxidases. Note that GH0 and GT0, denoted by gray with dotted lines, are not recognized enzyme families, but a bin for as-yet unclassified enzymes. Some of the smaller subfamilies are grouped together for ease of display
Supplementary table S1 shows similar abundance patterns for selected lignocellulose-degrading GH families across rain forest soil (this study), compost [1], cow rumen [7], and termite metagenome [48] datasets. GH family counts in the latter metagenomes were based on pfam hits to open reading frames on assembled metagenome contigs; however, the good correlation between BLASTX hits on unassembled reads and pfam hits on assembled ORFs suggests that BLASTX hits provide a reasonable proxy for enzyme family assignment. Beta-glucosidases and other oligosaccharide degrading families are the most abundant in the rain forest reads. The main cellulase families represented are GH5 (243 hits) and GH9 (86 hits), whereas other traditional cellulase families (GH7, GH45, GH48) are only present in very low abundance (if at all) in all four microbiomes examined.
Overall, the native soil community has a variety of potentially interesting genes for use in industrial biofuels production. However, the lack of assembly of full-length genes in this data set and the rather short list of full-length genes identified from compost in an earlier study by Allgaier et al. [1] prompted us to investigate whether selective cultivation on bioenergy feedstocks could reduce the community complexity of these native communities, thereby facilitating identification of a greater number of full-length genes in future metagenomic sequencing efforts.
Switchgrass-adapted Puerto Rican Rainforest Soil Cultures
We chose to adapt the tropical soil sample to switchgrass under anaerobic conditions to select for anaerobic biomass-degrading microbes, since many of these organisms produce cellulosomes, multi-enzyme complexes capable of depolymerizing both cellulose and hemicellulose. Anaerobic switchgrass-adapted consortia were enriched from tropical forest soils by passing the communities two times for 6 weeks, with switchgrass as the sole carbon source, under anaerobic conditions with and without supplemental iron (Table 1). The richness of the original soil sample was 1,339 taxa as determined by PhyloChip (Fig. 4a), and growth on switchgrass as the sole carbon source reduced the richness to 84 taxa, while inclusion of iron in the consortia growth media resulted in a richness of 336 taxa. There were archaea present in the soils that were not present in either feedstock-adapted community, along with taxa from 20 phyla (Table 2),
Microbial community profiles of native versus feedstock-adapted communities from b compost and a tropical forest soils. Profiles are based on the sequences of SSU rRNA genes using high-density PhyloChip microarray or SSU rRNA amplicon pyrosequencing. Richness of native communities is much higher compared to feedstock-adapted communities after growth on various feedstocks
Taxa in the switchgrass-adapted communities lacking iron were heavily dominated by Proteobacteria, Firmicutes, and Bacteroidetes (Fig. 4a), with members of the class Proteobacteria making up 83% of the richness of the switchgrass-adapted communities. All of the taxa enriched in the switchgrass-amended cultures lacking iron were also present in the iron-amended cultures (Fig. 4a). Taxa that were specifically enriched in the presence of iron and not found in the non-iron consortia were mostly dominated by the Bacteroidetes, Desulfovibrionaceae (class Deltaproteobacteria), Caulobacterales (class Alphaproteobacteria), and Enterococcales (class Bacilli). There were also representatives from many phyla that are rare, uncultivated, and otherwise of cryptic function (Table 2). Of the 41 phyla originally represented in the soil, only nine phyla remained when communities were adapted to switchgrass only, while iron addition resulted in the growth of taxa from 28 different phyla on switchgrass as the sole carbon source. These results clearly show that selective growth does indeed reduce the complexity of a native soil community (Table 3, Fig. 4).
Lignin-adapted Municipal Green Waste Compost Cultures
To test a different set of selective conditions, we adapted the municipal green waste compost community to various types of purified lignin as the sole carbon source under aerobic conditions, to select for microbes specialized in the deconstruction and/or modification of lignin (Table 1). The community was grown under aerobic conditions since many lignin-degrading enzymes use oxidative chemistry to depolymerize lignin. Three types of lignin were chosen as the carbon source: alkali lignin (AL), organosolv lignin (OL), and Indulin AT (IL). The AL is sulfonated and completely water-soluble with a MW range reported by Sigma Aldrich (St. Louis, MO) between 10,000 and 60,000. The OL is a mixture of soluble and insoluble lignin, indicating that it may have higher MW fragments of lignin than the AL. The IL is unsulfonated and completely insoluble (the soluble fraction was removed by water extraction), and therefore is likely to contain only high MW lignin. The range of water solubility and MW for these types of lignin is likely to facilitate the enrichment of microbes with a broad range of lignin-degrading attributes.
After five 2-week enrichments, the composition of each lignin-adapted microbial community was determined by SSU rRNA amplicon pyrosequencing. In the compost inoculum, a total of 391 different taxa were identified representing 24 bacterial and eukaryotic phyla. The diversity of the compost inoculum microbial community was reduced to 30, 98, and 136 taxa in the lignin-enriched cultures belonging to 15 bacterial phyla, with each of the three lignin-adapted cultures dominated by Alphaproteobacteria (Fig. 4b, Table 2). In general, eukaryotes were of minor abundance in both the compost inoculum and the lignin-adapted communities, with the exception of one taxon belonging to the Alveolata, which accounted for 2% of the microbial community in the OL enrichment.
Identifying the organisms shared between the three cultures may indicate which organisms are playing an active role in lignin modification and depolymerization. Eight phylotypes are common in all three lignin-amended cultures. Five of these phylotypes have cultured representatives: Paracoccus sp. Str. WB1, Mesorhizobium sp. Str. CCBAU 41182, brackish water isolate str. HINUF007, Rhizobium sp. str. RM1-2001, and Hyphomicrobium aestuarii str. DSM 1564. The other three phylotypes are most closely related to SSU rDNA clones recovered from environmental samples (water manure clone, aspen rhizosphere clone, and solid waste clone).
Again, the lignin-adapted communities showed a substantial decrease in microbial diversity compared to the native compost inoculum. The selective conditions were completely different from the switchgrass-adapted soil communities, yet both types of selection reduced the number of taxa compared to the respective native community, ranging from three- to 15-fold.
Discussion
Sequencing of individual genomes and metagenomes has advanced explosively in the last 5 years (e.g., Roche 454, Illumina, and Solexa). We can now sequence an organism's entire genome or an environmental metagenome in a few hours. Unfortunately, the resulting avalanche of bioinformatic data in terms of assembly and annotation has become a major bottleneck for utilizing this data. This is especially true for metagenomes from complex communities, where many previously unsequenced taxa are present [46]. Indeed, by most estimates, so far less than 1% of all microbial taxa have been identified, and less than 1% of those identified have been sequenced. Clearly, there is a vast, untapped genomic potential in many habitats waiting for the bioinformatic databases to catch up. Until that time, we need to devise ways to limit the complexity found in these untapped resources so that we can start to realize discovery of enzymes needed for enabling bioenergy breakthroughs. Examination of metagenomic datasets obtained from Puerto Rican rainforest soil indicates that this microbial community possesses many genes of interest to JBEI—such as cellulases, hemicellulases, and lignases—for deconstruction of feedstock biomass material, as well as enzymes and pathways for synthesis of new biofuels. Similar results were found earlier with municipal green waste compost [1]. The wide variety of enzyme families found in the rainforest soil (close to half of the 300 enzyme families described in CAZy and FOLy are present at five hits or more) is illustrative of the incredible diversity found in natural soil communities. Interestingly, about one sixth of the 16% of the sequence reads assigned by BLASTX to glycoside hydrolases match genes not yet assigned to a specific family (GH0 and GT0 in Fig. 3). This is a significantly higher fraction than was found in the compost sample (data not shown), suggesting that the rainforest environment may provide a rich source for as-yet uncharacterized families of glycoside hydrolases and glycosyl tranferases. However, despite the abundant sequence information, the shotgun sequence reads cannot be assembled into full-length enzymes because of the enormous complexity of soil microbial communities. Partial sequences of the length obtained in this study (averaging 417 bp) are too short to encapsulate many typical lignocellulolytic domains, let alone full-length genes. To obtain greater sequence coverage of the metagenome and genes of interest, our strategy is to simplify the microbial communities by generating specific feedstock-adapted consortia, which will possess the functional characteristics desirable for development of next-generation biofuels.
Since the tropical rain forest soil is rich in anaerobic microorganisms, we reduced the complexity of microbial community by growing them anaerobically on switchgrass as a sole carbon and energy source, and by enriching with iron as a terminal electron acceptor. By performing this selective enrichment, we were able to cultivate and characterize mixed microbial populations that are proficient in lignocellulosic degradation. The anaerobic enrichment yielded mostly Gammaproteobacteria from the family Enterobacteraceae. The Enterobacteriaceae were determined previously to play a strong role in anaerobic degradation of 13C-glucose, likely by mixed acid fermentation [15]. This group of facultative aerobes is most commonly identified with intestinal gut microbiota, though their role in anaerobic processes in soils is becoming more apparent [26]. Taxa from the groups Helicobacterales (Epsilonproteobacteria) and Pepttococcales (class Clostridia) are also generally facultative aerobes commonly associated with the gut microbial community, as pathogens as well as commensals [32]. Since the microbial community of ruminant animals is relatively well-characterized, and an active case study site for biofuels [48], it is encouraging to find classic cellulolytic taxa enriched in these communities. The anaerobic switchgrass-adapted tropical forest soil cultures also turned up Actinobacteria in the genus Beutenbergia sp., Arthrobacter sp., and Streptomyces. The Beutenbergia spp. were initially discovered as anaerobes isolated from a cave [22]. Arthrobacter spp. have so far only coincidentally been associated with lignocellulolytic environments, but enriched taxa are closely related to the taxa Streptomyces viridosporus, known to depolymerize lignin as it degrades lignocellulose via an extracellular lignin peroxidase [40].
Iron addition caused a large increase in the diversity of feedstock-adapted communities, likely because these cultures were limited for electron donors. These taxa were from many diverse phyla, but sulfate-reducing bacteria dominated, including members of the Deltaproteobacteria and Desulfotomaculum (phylum Firmicutes). There are myriad examples in the literature of sulfate-reducing bacteria (SRB) engaged in aromatic hydrocarbon degradation [20]. The role of SRBs in anaerobic decomposition of lignocellulose in natural environments has been well characterized for industrial applications, where leachate from paper pulp mills or municipal solid waste treatment plants seek to enrich SRBs for the purpose of bioremediation [16, 27]. Members of the family Desulfobacteriaceae have been shown to use cellulose fermentation products coupled with sulfidogenesis in co-cultures derived from a soda lake [52]. While it has not been shown directly in soils, it seems likely that the sulfate-reducing bacteria possess the ability to depolymerize and assimilate carbon from lignocellulose in the soil and couple it to sulfate reduction [41, 47]. The heretofore, poor characterization of sulfate-reducing bacteria in anaerobic lignocellulose degradation suggests opportunity for discovery of novel mechanisms of deconstruction.
The green waste compost community, which is dominated by aerobic microorganisms, was reduced in complexity by growing the community on lignin as a sole carbon source under aerobic conditions. Lignin-adapted cultures were also enriched for a variety of organisms represented by different phyla. The most abundant organism in the AL-amended culture was a member of the phylum Actinobacteria, related to Rhodococcus sp. Isolates of the genus Rhodococcus have been shown to degrade a wide variety of aromatic compounds, hydrophobic natural compounds, and xenobiotics, including lignin-like compounds, and may be responsible for lignin degradation in the AL culture [2, 3, 13, 17]. One organism present in all three lignin-amended cultures is related to the brackish water isolate str. HINUF007. Its SSU rDNA sequence is related to the family Sphingomonadaceae (98.9% identity to Sphingomonadaceae bacterium clone IWENVB15). Sphingomonas paucimobilis has been extensively characterized for its role in lignin degradation and can catabolize several lignin-derived biaryls and monoaryls [35]. Therefore, the Sphingomonas-related organism in the lignin-amended cultures may have an important role in lignin degradation. Other phyla represented in these lignin-adapted cultures include Acidobacteria and Verrucomicrobia. Little is known about these phyla, yet both are prevalent in soil and are proposed to deconstruct complex organic matter [29, 38]. Isolation of lignin-degrading members of these phyla or genome reconstruction of these members from metagenomic sequencing data will provide important insights into their ability to degrade lignin.
The SSU rRNA community profiles also indicate that a second unintentional selection force may be at work in the lignin-amended cultures. Four of the phylotypes shared between the three cultures (Paracoccus, Mesorhizobium, Rhizobium, and Hyphomicrobium) have cultured representatives with the ability to perform denitrification under low or fluctuating oxygen concentrations [4, 18, 19]. The primary nitrogen source in the lignin-amended cultures is NH +4 ; however, the trace elements solution used to supply essential metals contain the chelator nitrilotriacetate (NTA). Nitrilotriacetate is a biodegradable carbon source and many NTA-utilizing bacteria have been characterized [8]. Aerobic microorganisms initiate NTA degradation with the enzyme NTA monooxygenase, a gene present in both Mesorhizobium and Paraccocus genomes as determined from a search of NCBI [8]. Although the concentration of NTA in the cultures was low (about 0.5 µM), it is unclear whether these organisms were consuming NTA or lignin. Removal of NTA from future enrichment cultures should clarify this issue.
The exponential advances that have occurred in pyrosequencing in just the last 5 years have made us appreciate even more “The Doctrine of Infallibility”— i.e., there is no compound, man-made or natural, that microorganisms cannot degrade. However, until we sequence more of the extant microbial community and develop better and higher throughput techniques for genome assembly and annotation, we need to reduce community complexity in ways that are appropriate for discovering new, essential enzymes. This is especially true for discovering lignocellulosic enzymes that can be used for production of alternative liquid fuels. Choosing the appropriate environmental community and enrichment conditions, (e.g., anaerobic or aerobic) is necessary in selecting for microbes that can deconstruct lignocellulosic biomass under industrial pretreatment-relevant conditions. We demonstrated that metagenome and SSU rRNA genotyping of soil and compost microbial communities show they are inhabited by lignocellulose-degrading microbes, and are thus good candidates for starting inocula. The decrease in complexity of these communities will likely increase sequence coverage of the metagenome using 454 pyrosequencing, increasing the number of full-length genes identified, and facilitating the discovery of lignocellulose-degrading enzymes. Selective community enrichments like these should greatly facilitate our ability to use the latest advances in genomic sequencing as a platform for high throughput discovery of unique new enzymes, which will advance the development of lignocellulosic liquid biofuels.
References
Allgaier M, Reddy A, Park JI, Ivanova N, D'Haeseleer P, Lowry S et al (2010) Targeted discovery of glycoside hydrolases from a switchgrass-adapted compost community. Plos One 5:9
Andreoni V, Bernasconi S, Bestetti P, Villa M (1991) Metabolism of lignin-related compounds by Rhodococcus rhodochorous-bioconversion of anisoin. Appl Microbiol Biotechnol 36:410–415
Barnes MR, Duetz WA, Williams PA (1997) A 3-(3-hydroxyphenyl)propionic acid catabolic pathway in Rhodococcus globerulus PWD1: Cloning and characterization of the hpp operon. J Bacteriol 179:6145–6153
Baumann B, Snozzi M, Zehnder AJ, Van Der Meer JR (1996) Dynamics of denitrification activity of Paracoccus denitrificans in continuous culture during aerobic-anaerobic changes. J Bacteriol 178:4367–4374
Braid MD, Daniels LM, Kitts CL (2003) Removal of PCR inhibitors from soil DNA by chemical flocculation. J Microbiol Meth 52:389–393
Brodie EL, DeSantis TZ, Joyner DC, Baek SM, Larsen JT, Andersen GL et al (2006) Application of a high-density oligonucleotide microarray approach to study bacterial population dynamics during uranium reduction and reoxidation. Appl Environ Microbiol 72:6288–6298
Brulc JM, Antonopoulos DA, Miller MEB, Wilson MK, Yannarell AC, Dinsdale EA et al (2009) Gene-centric metagenomics of the fiber-adherent bovine rumen microbiome reveals forage specific glycoside hydrolases. Proc Natl Acad Sci USA 106:1948–1953
Bucheli-Witschel M, Egli T (2001) Environmental fate and microbial degradation of aminopolycarboxylic acids. FEMS Microbiol Rev 25:69–106
Cantarel BL, Coutinho PM, Rancurel C, Bernard T, Lombard V, Henrissat B (2009) The Carbohydrate-Active EnZymes database (CAZy): an expert resource for Glycogenomics. Nucleic Acids Res 37:D233–D238
Charles D (2009) Corn-based ethanol flunks key test. Science 324:587–587
Chou HH, Holmes MH (2001) DNA sequence quality trimming and vector removal. Bioinformatics 17:1093–1104
Claudel-Renard C, Chevalet C, Faraut T, Kahn D (2003) Enzyme-specific profiles for genome annotation: PRIAM. Nucleic Acids Res 31:6633–6639
de Carvalho C, da Fonseca MMR (2005) The remarkable Rhodococcus erythropolis. Appl Microbiol Biotechnol 67:715–726
DeAngelis KM, Silver WL, Thompson AW, Firestone MK (2010) Microbial communities acclimate to recurring changes to soil redox potential status. Environ Microbiol, in press
Degelmann DM, Kolb S, Dumont M, Murrell JC, Drake HL (2009) Enterobacteriaceae facilitate the anaerobic degradation of glucose by a forest soil. FEMS Microbiol Ecol 68:312–319
Detmers J, Schulte U, Strauss H, Kuever J (2001) Sulfate reduction at a lignite seam: microbial abundance and activity. Microb Ecol 42:238–247
Eulberg D, Golovleva LA, Schlomann M (1997) Characterization of catechol catabolic genes from Rhodococcus erythropolis 1CP. J Bacteriol 179:370–381
Fesefeldt A, Kloos K, Bothe H, Lemmer H, Gliesche CG (1998) Distribution of denitrification and nitrogen fixation genes in Hyphomicrobium spp. and other budding bacteria. Can J Microbiol 44:181–186
Garcia-Plazaola JI, Becerril JM, Arreseigor C, Hernandez A, Gonzalezmurua C, Apariciotejo PM (1993) Denitrifying ability of 13 Rhizobium meliloti strains. Plant Soil 149:43–50
Gibson J, Harwood CS (2002) Metabolic diversity in aromatic compound utilization by anaerobic microbes. Annu Rev Microbiol 56:345–369
Giroux H, Vidal P, Bouchard J, Lamy F (1988) Degradation of kraft indulin by Streptomyces viridosporus and Streptomyces badius. Appl Environ Microbiol 54:3064–3070
Groth I, Schumann P, Schuetze B, Augsten K, Kramer I, Stackebrandt E (1999) Beutenbergia cavernae gen. nov., sp, nov., an l-lysine-containing actinomycete isolated from a cave. Int J Syst Bacteriol 49:1733–1740
Hershberger KL, Barns SM, Reysenbach AL, Dawson SC, Pace NR (1996) Wide diversity of Crenarchaeota. Nature 384:420–420
Jaeger KE, Eggert T (2002) Lipases for biotechnology. Curr Opin Biotechnol 13:390–397
Janssen PH, Schuhmann A, Morschel E, Rainey FA (1997) Novel anaerobic ultramicrobacteria belonging to the Verrucomicrobiales lineage of bacterial descent isolated by dilution culture from anoxic rice paddy soil. Appl Environ Microbiol 63:1382–1388
Kersters K (2006) The Genus Deleya. Prokaryotes: a handbook on the biology of bacteria, vol. 6, 3rd edn: Proteobacteria: Gamma Subclass, 836-843
Kim J, Kim M, Bae W (2009) Effect of oxidized leachate on degradation of lignin by sulfate-reducing bacteria. Waste Manag Res 27:520–526
Kunin WE, Vergeer P, Kenta T, Davey MP, Burke T, Woodward FI et al (2009) Variation at range margins across multiple spatial scales: environmental temperature, population genetics and metabolomic phenotype. Proceedings of the Royal Society B-Biological Sciences 276:1495–1506
Lee KC, Webb RI, Janssen PH, Sangwan P, Romeo T, Staley JT, et al. (2009) Phylum Verrucomicrobia representatives share a compartmentalized cell plan with members of bacterial phylum Planctomycetes. BMC Microbiology 9
Lee SK, Chou H, Ham TS, Lee TS, Keasling JD (2008) Metabolic engineering of microorganisms for biofuels production: from bugs to synthetic biology to fuels. Curr Opin Biotechnol 19:556–563
Levasseur A, Plumi F, Coutinho PM, Rancurel C, Asther M, Delattre M et al (2008) FOLy: an integrated database for the classification and functional annotation of fungal oxidoreductases potentially involved in the degradation of lignin and related aromatic compounds. Fungal Genet Biol 45:638–645
Ley RE, Hamady M, Lozupone C, Turnbaugh PJ, Ramey RR, Bircher JS et al (2008) Evolution of mammals and their gut microbes. Science 320:1647–1651
Li LL, McCorkle SR, Monchy S, Taghavi S, van der Lelie D (2009) Bioprospecting metagenomes: glycosyl hydrolases for converting biomass. Biotechnology for Biofuels 2:11
Lopez MJ, Vargas-Garcìa MdC, Suarez-Estrella F, Nichols NN, Dien BS, Moreno J (2007) Lignocellulose-degrading enzymes produced by the ascomycete Coniochaeta ligniaria and related species: application for a lignocellulosic substrate treatment. Enzyme Microb Technol 40:794–800
Masai E, Katayama Y, Fukuda M (2007) Genetic and biochemical investigations on bacterial catabolic pathways for lignin-derived aromatic compounds. Biosci Biotechnol Biochem 71:1–15
McGroddy M & Silver WL (2000) Variations in belowground carbon storage and soil CO2 flux rates along a wet tropical climate gradient. Vol. 32 ed.^eds, pp. 614-624
Meyer F, Paarmann D, D'Souza M, Olson R, Glass EM, Kubal M et al (2008) The metagenomics RAST server—a public resource for the automatic phylogenetic and functional analysis of metagenomes. BMC Bioinformatics 9:8
Mou XZ, Sun SL, Edwards RA, Hodson RE, Moran MA (2008) Bacterial carbon processing by generalist species in the coastal ocean. Nature 451:708–U704
Parton W, Silver WL, Burke IC, Grassens L (2007) Global-scale similarities in nitrogen release patterns during long-term decomposition. Science 315:361–4
Pasti MB, Pometto AL, Nuti MP, Crawford DL (1990) Lignin-solubilizing ability of Actinomycetes isolated from termite (Termitidae) hindgut. Appl Environ Microbiol 56:2213–2218
Seeliger S, Cord-Ruwisch R, Schink B (1998) A periplasmic and extracellular c-type cytochrome of Geobacter sulfurreducens acts as a ferric iron reductase and as an electron carrier to other acceptors or to partner bacteria. J Bacteriol 180:3686–3691
Silver WL, Lugo AE, Keller M (1999) Soil oxygen availability and biogeochemistry along rainfall and topographic gradients in upland wet tropical forest soils. Biogeochemistry 44:301–328
Tanner RS (2007) Cultivation of bacteria and fungi. Manual of Environmental Microbiology 69-78
Tschech A, Pfennig N (1984) Growth-yield increase linked to caffeate reduction in Acetobacterium woodii. Arch Microbiol 137:163–167
Turnbaugh PJ, Hamady M, Yatsunenko T, Cantarel BL, Duncan A, Ley RE et al (2009) A core gut microbiome in obese and lean twins. Nature 457:480–U487
Vogel TM, Simonet P, Jansson JK, Hirsch PR, Tiedje JM, van Elsas JD et al (2009) TerraGenome: a consortium for the sequencing of a soil metagenome. Nat Rev Microbiol 7:252–252
Walker CB, He ZL, Yang ZK, Ringbauer JA, He Q, Zhou JH et al (2009) The electron transfer system of syntrophically grown Desulfovibrio vulgaris. J Bacteriol 191:5793–5801
Warnecke F, Luginbuhl P, Ivanova N, Ghassemian M, Richardson TH, Stege JT et al (2007) Metagenomic and functional analysis of hindgut microbiota of a wood-feeding higher termite. Nature 450:560–U517
Weaver PL, Murphy PG (1990) Forest structure and productivity in Puerto Rico Luquillo Mountains. Biotropica 22:69–82
Widdel F, Kohring GW, Mayer F (1983) Studies on dissimilatory sulfate-reducing bacteria that decompose fatty-acids. 3. Characterization of the Filamentous Gliding Desulfonema limicola gen-nov sp-nov, and Desulfonema magnum sp-nov. Arch Microbiol 134:286–294
Wilson KH, Blitchington RB, Greene RC (1990) Amplification of bacterial-16s ribosomal DNA with polymerase chain-reaction. J Clin Microbiol 28:1942–1946
Zavarzin GA, Zhilina TN, Dulov LE (2008) Alkaliphilic sulfidogenesis on cellulose by combined cultures. Microbiology 77:419–429
Acknowledgements
The authors would like to especially thank Dr. Ken Vogel of the USDA for the switchgrass samples used in this study. This work was part of the DOE Joint BioEnergy Institute (http://www.jbei.org) supported by the US Department of Energy, Office of Science, Office of Biological and Environmental Research, through contract DE-AC02-05CH11231 between Lawrence Berkeley National Laboratory and the US Department of Energy.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Author information
Authors and Affiliations
Corresponding author
Additional information
Kristen M. DeAngelis and John M. Gladden contributed equally to this manuscript.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Table S1
Inventory of selected lignocellulose-degrading glycoside hydrolase families (GHs) identified in the rain forest soil microbiome. (DOC 353 kb)
Rights and permissions
Open Access This is an open access article distributed under the terms of the Creative Commons Attribution Noncommercial License (https://creativecommons.org/licenses/by-nc/2.0), which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
About this article
Cite this article
DeAngelis, K.M., Gladden, J.M., Allgaier, M. et al. Strategies for Enhancing the Effectiveness of Metagenomic-based Enzyme Discovery in Lignocellulolytic Microbial Communities. Bioenerg. Res. 3, 146–158 (2010). https://doi.org/10.1007/s12155-010-9089-z
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12155-010-9089-z