Abstract
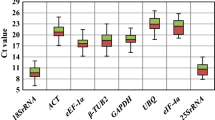
Due to its accuracy, sensitivity and reproducibility, real-time PCR has emerged as a key technique to measure changes in expression of target genes. But to obtain reliable results using real-time PCR, gene transcripts that do not alter its expression under different conditions need to be identified. These transcripts can then be used as references to normalize the expression of target genes. Only a few studies have been conducted so far in the identification of such reference genes in plants. In an effort to identify appropriate reference genes, cassava, an important food source in the tropics and an excellent crop against famine and drought, was assessed for its capacity to express eight housekeeping genes in leaves, stems and roots at four different stages of development under field conditions. The eight candidate genes tested were rRNA18S(18S), β-tubulin(TUB), actin 11(ACT), elongation factor 1α(EF1), translation initiation factor 5A(F5A), ubiquitin protein ligase E3-2a(UBI), ubiquitin conjugating enzyme E2-10(U10) and ubiquitin conjugating enzyme E2-35(U35). Data analysis was performed using four approaches commonlyemployed in identifying the reference genes and consensus rankings were generated using the output of each independent approach. EF1 and TUB were the most stable genes when all tissues and developmental stages were analyzed together. At different stages of development, UBI/18S, EF1/U35, TUB/U35werethe most stable genes in roots, leaves and stem tissues, respectively. Our results suggest the use of more than one reference gene in gene expression studies involving different tissues at different developmental stages.






Similar content being viewed by others
Abbreviations
- ACT:
-
Actin 11 gene
- DAP:
-
Days after planting
- EF1:
-
Elongation factor 1α gene
- F5A:
-
Translation initiation factor 5A gene
- HKG:
-
Housekeeping genes
- SD:
-
Standard deviation
- TUB:
-
β-tubulin gene
- U10:
-
Ubiquitin conjugating enzyme E2-10 gene
- U35:
-
Ubiquitin conjugating enzyme E2-35gene
- UBI:
-
Ubiquitin protein ligase E3-2a gene
- 18S:
-
rRNA18S gene
References
Alves CA (2002) Cassava botany and physiology. In: Hillocks RJ, Thresh JM, Bellotti AC (eds) Cassava: biology, production and utilization. CABI, New York, pp 67–90
Andersen C, Jensen J, Ørntoft T (2004) Normalization of real-time quantitative reverse transcription PCR data: a model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Res 64:5245–5250
Artico S, Nardeli SM, Brilhante O et al (2010) Identification and evaluation of new reference genes in Gossypiumhirsutum for accurate normalization of real-time quantitative RT-PCR data. BMC Plant Biol 10:49–61
Beekman L, Tohver T, Dardari R et al (2011) Evaluation of suitable reference genes for gene expression studies in bronchoalveolar lavage cells from horses with inflammatory airway disease. BMC Mol Biol 12:5–15
Beltrán J, Jaimes H, Echeverry M et al (2008) Quantitative analysis of transgenes in cassava plants using real-time PCR technology. In Vitro Cell Dev Biol Plant 45:48–56
Beltrán J, Prías M, Al-Babili S et al (2010) Expression pattern conferred by a glutamic acid-rich protein gene promoter in weld-grown transgenic cassava (ManihotesculentaCrantz). Planta 231:1413–1424
Bull SE, Owiti JA, Niklaus M et al (2009) Agrobacterium-mediated transformation of friable embryogeniccalli and regeneration of transgenic cassava. Nat Protoc 4:1845–1854
Czechowski T, Stitt M, Altmann T et al (2005) Genome-wide identification and testing of superior referencegenes for transcript normalization in Arabidopsis. Plant Physiol 139:5–17
Duarte Silveira É, Alves-Ferreira M, Guimarães L et al (2009) Selection of reference genes for quantitative real-time PCR expression studies in the apomictic and sexual grass Brachiariabrizantha. BMC Plant Biol 9:84–93
Echeverry M (2008) Quantification of transcriptional activity of genes associated with cyanogenesis in cassava (ManihotesculentaCrantz). Dissertation. University of Puerto Rico, Mayaguez
Echeverry M, Ocasio-Ramirez V, Figueroa A et al (2013) Expression profiling of genes associated with cyanogenesis in three cassava cultivars containing varying levels of toxic cyanogens. Amer J Plant Sci 4:1533–1545
Eggermont K, Goderis IJ, Broekaert WF (1996) High-throughput RNAextraction from plant samples based on homogenization by reciprocal shaking in the presence of a mixture of sand and glass beads. Plant Mol Biol Rep 14:273–279
Eggum BO (1970) The protein quality of cassava leaves. Brit J Nutr 24:761–768
El-Sharkawy MA (2004) Cassava biology and physiology. Plant Mol Biol 56:481–501
Graeber K, Linkies A, Wood A et al (2011) A guideline to family-wide comparative state-of-the-art quantitative RT-PCR analysis exemplified with aBrassicaceaecross-species seed germination case study. Plant Cell 23:2045–2063
Halsey ME, Olsen KM, Taylor NJ et al (2008) Reproductive biology of cassava (ManihotesculentaCrantz). Crop Sci 48:49–58
He JQ, Sandford AJ, Wang IM et al (2008) Election of housekeeping genes for real-time PCR in atopic human bronchial epithelial cells. Eur Respir J 32:755–762
Ihemere U, Arias-Garzon D, Lawrence S et al (2006) Genetic modification of cassava for enhanced starch production. Plant Biotechnol J4:453–465
Jain M, Nijhawan A, Tyagi AK et al (2006) Validation of housekeeping genes as internal control for studying gene expression in rice by quantitative real-time PCR. Biochem Biophys Res Commun 345:646–651
Jørgensen K, Morant A, Morant M et al (2005) Cassava plants with a depletedcyanogenicglucosidecontent in leaves and tubers. Distribution of cyanogenicglucosides, their site of synthesis and transport, and blockage of the biosynthesis by RNA interference technology. Plant Physiol 139:363–374
Lancaster P, Brooks J (1983) Cassava leaves as human food. Econ Bot 37:331–348
Li QF, Sun SSM, Yuan DY et al (2010) Validation of candidate reference genes for the accurate normalization of real-time quantitative RT-PCR data in rice during seed development. Plant Mol Biol Rep 28:49–57
Liu J, Zheng Q, Ma Q et al (2011) Cassava genetic transformation and its application in breeding. J Integr Plant Biol 53:552–569
Lopez C, Soto M, Restrepo S et al (2005) Gene expression profile in response to Xanthomonasaxonopodispv. manihotis infection in cassava using a cDNA microarray. Plant Mol Biol 57:393–410
Mallona I, Lischewski S, Weiss J et al (2010) Validation of reference genes for quantitative real-time PCR during leaf and flower development in Petunia hybrid. BMC Plant Biol 10:4–15
Nassar NMA, Marques AO (2006) Cassava leaves as a source of protein. J Agric Environ 4:187–189
Nicot N, Hausman JF, Hoffmann L et al (2005) Housekeeping gene selection for real-time RT-PCR normalization in potato during biotic and abiotic stress. J Exp Bot 56:2907–2914
Patil B, Ogwok E, Wagaba H et al (2011) RNAi-mediated resistance to diverse isolates belonging to two virus species involved in cassava brown streak disease. Mol Plant Path 12:31–41
Ogwok E, Odipio J, Halsey M et al (2012) Transgenic RNA interference (RNAi)-derived field resistance to cassava brown streak disease. Mol Plant Path 13:1019–1031
Ojulong H, Labuschangne MT, Fregene M et al (2008) A cassava clonal evaluation trial based on new cassava breeding scheme. Euphytica 160:119–129
Pfaffl MW (2001) A new mathematical model for relative quantification in real time RT-PCR. Nucleic Acids Res 29:9e45
Pfaffl MW, Tichopad A, Prgomet C et al (2004) Determination of stable housekeeping genes, differentially regulated target genes and sample integrity: Bestkeeper –Excel-based tool using pair-wise correlations. Biotechnol Lett 26:509–515
Prochnik S, Reddy Marri P, Desany B et al (2012) The cassava genome: current progress, future directions. Trop Plant Biol 5:88–94
Putten H, Sudarmonowati E, Herman M et al (2012) Field testing and exploitation of genetically modified cassava with low-amylose or amylose-free starch in Indonesia. Transgenic Res 21:39–50
Sayre R, Beeching JR, Cahoon EB et al (2011) The BioCassavaPlus Program: biofortification of cassava for sub-saharan Africa. Ann Rev Plant Biol 62:251–272
Silver N, Best S, Jiang J et al (2006) Selection of housekeeping genes for gene expression studies in human reticulocytes using real-time PCR. BMC Mol Biol 7:33–42
Siritunga D, Sayre R (2003) Generation of cyanogen-free transgenic cassava. Planta 217:367–373
Siritunga D, Sayre R (2004) Engineering cyanogen synthesis andturnover in cassava (Manihot esculenta). Plant Mol Biol 56:661–669
Siritunga D, Arias-Garzon D, White W et al (2004) Over-expression of hydroxyl nitrilelyase in transgenic cassava roots accelerates cyanogenesis and food detoxification. Plant Biotechnol J 2:37–43
Ståhlberg A, Elbing K, Andrade-Garda JM et al (2008) Multiwayreal-time PCR gene expression profiling yeast Saccharomyces cerevisiae reveals altered transcriptional response of ADH-genes to glucose stimuli. BMC Genomics 9:170–184
Stern-Straeter J, Bonaterra GA, Hörmann K et al (2009) Identification of valid reference genes during the differentiation of human myoblasts. BMC Mol Biol 10:66–74
Suslov O, Steindler D (2005) PCR inhibition by reverse transcriptase leads to an overestimation of amplification efficiency. Nucleic Acids Res 33:20e181
Taylor NJ, Halsey M, Gaitán-Solís E et al (2012a) The VIRCA Project: virus resistant cassava for Africa. GM Crops Food 3:93–103
Taylor NJ, Gaiton-Solis E, Moll T et al (2012b) A high-throughput platform for theproduction and analysis of transgeniccassava (Manihotesculenta) Plants. Trop Plant Biol 5:127–139. doi:10.1007/s12042-012-9099-4
Utsumi Y, Tanaka M, Morosawa T et al (2012) Transcriptomeanalysis using a high-density oligomicroarray under drought stress in various genotypes of cassava: an Important Tropical Crop. DNA Res 19:335–345
Vanderschuren H, Moreno I, Anjanappa RB et al (2012) Exploiting the combination of natural and genetically engineered resistance to cassava mosaic and cassava brown streak viruses impacting cassava production in Africa. PLoS ONE 7:9, e45277
Vandesompele J, De Preter K, Pattyn F et al. (2002) Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol 3:1–12
Welsch R, Arango J, Bär C, Salazar B et al (2010) Provitamin A accumulation in cassava (Manihot esculenta) roots driven by a single nucleotide polymorphism in a phytoene synthase gene. Plant Cell 22:3348–3356
Yadav J, Ogwok E, Wagaba H et al (2011) RNAi-mediatedresistance to cassava brown streak Uganda virus in transgenic cassava. Mol Plant Pathol 12:677–687
Zainuddin I, Schlegel K, Gruissem W et al (2012) Robust transformation procedure for the production of transgenic farmer-preferred cassava landraces. Plant Methods 8:24–31
Zhao S, Fernald RD (2005) Comprehensive algorithm for quantitative real-time polymerase chain reaction. J Comput Biol 12:1047–1064
Zidenga T, Leyva-Guerrero E, Moon H et al (2012) Extending cassava root shelf life via reduction of reactive oxygen species production. Plant Physiol 159:1396–1407
Zhang P, Vanderschuren H, Futterer J et al (2005) Resistance to cassava mosaic disease in transgenic cassava expressing antisense RNAs targeting virus replication genes. Plant Biotechnol J 3:385–389
Acknowledgments
We would like to express our gratitude to Pedro Marquez, Director of the Isabela Agricultural Research Station (in Isabela, PR), and the field crew for proper maintenance of cassava plants. We also thank Dr. John Uscian (Department of Biology, University of Puerto Rico Mayaguez, Mayaguez, PR) for the critical review of this manuscript.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by: Paulo Arruda
Electronic supplementary material
Below is the link to the electronic supplementary material.
ESM 1
(DOCX 20 kb)
Rights and permissions
About this article
Cite this article
Salcedo, A., Zambrana, C. & Siritunga, D. Comparative Expression Analysis of Reference Genes in Field-Grown Cassava. Tropical Plant Biol. 7, 53–64 (2014). https://doi.org/10.1007/s12042-014-9137-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12042-014-9137-5