Abstract
Somatic embryogenesis can be used for large-scale propagation of plants. In most conifers, it is only possible to establish embryogenic cultures from zygotic embryos or young seedlings. There is, however, a great interest to propagate selected trees with valuable traits via somatic embryos. To be able to establish embryogenic cultures from adult tissues, more knowledge about the molecular regulation of totipotency and embryogenic potential is needed. In Arabidopsis (Arabidopsis thaliana), LEAFY COTYLEDON1 (LEC1) is required for somatic embryogenesis, and overexpression of LEC1 can stimulate the formation of embryo-like structures from vegetative tissues. We have previously characterized a conifer LEC1-type gene, PaHAP3A, which is preferentially active during embryo development in Norway spruce (Picea abies). In this work, we show, by using histochemical GUS assays, that PaHAP3A is expressed in germinated embryos at presumptive sites from which embryogenic tissue differentiate during initiation of embryogenic cultures. Furthermore, we have overexpressed PaHAP3A, using both constitutive and inducible promoters, in order to elucidate whether elevated transcript levels of PaHAP3A are sufficient to induce embryonic properties after germination. In contrast to its angiosperm counterpart, PaHAP3A does not stimulate embryonic features in vegetative tissues. However, overexpression of PaHAP3A during the maturation stage leads to the differentiation of ectopic embryos from maturing somatic embryos. Our results not only indicate at least partial conservation between the conifer and angiosperm LEC1-type genes but also suggest that PaHAP3A is dependent of the cellular status to confer its presumptive role as an important factor influencing the competence of the tissue to initiate somatic embryos.
Similar content being viewed by others
Introduction
During their life cycle, higher plants undergo distinct transitions through embryonal, vegetative, and reproductive developmental stages. The life cycle starts with the fertilization of egg cells by sperm cells to produce zygotic embryos. The embryonal stage can be divided into two distinct phases, an early morphogenic phase and a late maturation phase (West 1993; Goldberg et al. 1994). Seed germination marks the end of the embryonal development, and rapid repression of embryonic genes is observed with seed imbibition (Tai et al. 2005). Developmental transitions require the precise temporal and spatial expression of regulatory genes, modulated by epigenetic mechanisms. Histone deacetylases (HDACs) are involved in the repression of embryonic properties upon germination, and inhibition of HDAC activity results in growth arrest and the formation of embryo-like structures after germination, possibly through spurious expression of embryo-specific factors (Tanaka et al. 2007). A variety of plant cells have been demonstrated to possess the capacity to redirect development and regain their totipotent state, a characteristic that can be manifested in the capacity to differentiate into somatic embryos. Initiation of somatic embryogenesis requires an induction signal that causes a somatic cell to change its identity and become embryogenic and also that the cell is competent to respond to the signal. Phytohormones play an important role during zygotic and somatic embryogenesis, and auxin is suggested to be the universal induction signal for somatic embryogenesis (Braybrook and Harada 2008). In addition to auxin being necessary for the initiation of somatic embryogenesis in most plant species, it also plays a central role during embryo development (reviewed in e.g., ten Hove et al. 2015). High levels of abscisic acid (ABA) and low levels of gibberellic acid (GA) are necessary for embryo maturation, whereas the ratio is reversed during embryo germination (reviewed in Gutierrez et al. 2007).
Knowledge of the genetic regulation of plant embryogenesis has to a large extent been obtained by studying gain- and loss-of-function mutants in the angiosperm model plant Arabidopsis (Arabidopsis thaliana). Several genes have been shown to stimulate the formation of embryogenic callus or somatic embryos from vegetative cells. Expression of Somatic Embryogenesis Receptor-like Kinase1 (SERK1) marks embryogenic competence, but the gene can also be expressed in non-embryogenic cells (Schmidt et al. 1997; Hecht et al. 2001). Ectopic expression of BABY BOOM (BBM) primarily induces spontaneous somatic embryo formation in seedlings (Boutilier et al. 2002). Overexpression of WUSCHEL (WUS) promotes the formation of somatic embryos from various vegetative tissues without an extra supply of plant hormones (Zuo et al. 2002). Ectopic expression of AGAMOUS-Like 15 (AGL15) affects the embryonic program and enhances the competence of a shoot apical meristem to undergo somatic embryogenesis (Harding et al. 2003). LEAFY COTYLEDON1 (LEC1) is an important regulator of both early and late embryogenesis and is also required for somatic embryogenesis (Gaj et al. 2005; Braybrook and Harada 2008). LEC1, which encodes a HAP3 subunit of the CCAAT-BOX BINDING FACTOR (CBF), is specifically expressed in the seed, both in the embryo and the endosperm, and its expression becomes repressed during the switch from embryonic to vegetative growth (Lotan et al. 1998). Ectopic expression of LEC1 in Arabidopsis induces a range of abnormalities. Root and hypocotyl growth of seedlings are impaired, secondary cotyledons develop instead of leaves, and embryo-like structures emerge from vegetative tissue and express embryo-specific genes that encode seed storage proteins (Lotan et al. 1998). Another gene encoding a LEC1-type HAP3 subunit in Arabidopsis, LEC1-like (L1L), is required for embryogenesis but with distinct roles from LEC1 (Kwong et al. 2003). Nevertheless, ectopic expression of L1L can rescue the defects of the lec1 mutant. PICKLE (PKL), which encodes a CHD3 chromatin-remodeling factor, is required for the repression of LEC1 and other embryo master regulators during and after germination (Ogas et al. 1997; Ogas et al. 1999; Rider et al. 2003).
We have previously identified two LEC1-type genes in Norway spruce (Picea abies), PaHAP3A and PaHAP3B, and a comparative phylogenetic analysis of HAP3 genes suggested that PaHAP3A and PaHAP3B are paralogous genes originating from a duplication event in the conifer lineage (Uddenberg et al. 2011). PaHAP3A is a conifer homolog to LEC1 and L1L. The expression of PaHAP3A is high during early embryo development but decreases during late embryogeny. Chemical treatment of maturating somatic embryos of Norway spruce with the HDAC inhibitor trichostatin A (TSA) arrests maturation simultaneously as the expression of PaHAP3A remains high. TSA treatment also alters the expression of another embryogenesis-related gene, the Norway spruce homologue to ABSCISIC ACID INSENSITIVE3/VIVIPAROUS1, PaVP1. In addition, TSA treatment of germinating embryos of Norway spruce partially inhibits the progression of germination and maintains the embryogenic potential (Uddenberg et al. 2011).
In this work, we have asked the questions if overexpression of PaHAP3A stimulates the embryogenic potential and if it affects embryo maturation and germination in a similar way as TSA treatment. Our results indicate that the PaHAP3A expression coincides with early embryogenesis and localizes to foci on germinated embryo explants from which new embryogenic tissue differentiate. Overexpression of PaHAP3A is neither sufficient to affect normal progression of embryo maturation or germination nor sufficient to alter the embryogenic potential after germination. This shows that TSA has a wider effect on gene expression patterns than that governed by PaHAP3A. Nevertheless, increased activity of the conifer LEC1-type gene during embryo maturation is sufficient for ectopic embryo formation.
Materials and methods
Plant material and growth conditions
Embryogenic cell lines 88.22 and 61.21 of Norway spruce (Picea abies L. Karst.) were used in this study. Cell line 88.22 was chosen since this cell line was used in our previous study of the expression of PaHAP3A during embryo development and germination (Uddenberg et al. 2011), and cell line 61.21 because this is the cell line that we routinely use for studying gene function in Norway spruce (e.g., Alvarez et al. 2015). Embryogenic cultures were treated as previously described (von Arnold and Clapham 2008), except that the cultures were proliferated on solidified medium. Briefly, the cultures were proliferated as proembryogenic masses (PEMs) on proliferation medium containing the plant growth regulators (PGRs), auxin (2,4-dichlorophenoxyacetic acid), and cytokinin (N6-benzyladenine). To stimulate differentiation of early somatic embryos from PEMs, the cultures were transferred to prematuration medium, i.e., medium lacking PGRs, for 1 week. Further development of the embryos was stimulated by transferring the cultures to maturation medium containing abscisic acid (ABA). Late embryos developed within 2 weeks on maturation medium, and mature embryos were formed after 4–8 weeks of maturation treatment. Mature embryos were desiccated for 1 week before they were transferred to germination medium (von Arnold and Clapham 2008).
RNA extraction, cDNA synthesis, and quantitative real-time PCR
Total RNA from embryogenic cultures was isolated using the Spectrum Plant Total RNA kit (Sigma-Aldrich), according to the manufacturer’s instruction. Isolated RNA was subjected to DNase digestion using the accessory on-column DNase digestion kit (Sigma-Aldrich). Complementary DNA (cDNA) was prepared from 1-μg total RNA using the Maxima First Strand cDNA Synthesis Kit for qPCR (Thermo Scientific).
Quantitative real-time PCR (qPCR) was performed using an iQ5 Real-Time Detection System in iCycler iQ 96-well PCR plates with adhesive seals (Bio-Rad). Primers used to quantify expression levels are presented in Table S1. Target gene expression values were normalized against the reference genes PHOSPHOGLUCOMUTASE (PaPHOS), ELONGATION FACTOR1-α (PaEF1-α), and CELL DIVISION CYCLE 2 (PaCDC2), previously selected based on their stability during embryo development (Vestman et al. 2011) using the geNorm software (Vandesompele et al. 2002). Amplifications were carried out using the DyNAmo Flash Sybr Green qPCR kit (Thermo Scientific). PCR cycling conditions were as advised by the manufacturer, with annealing and extension at 60 °C for 30 s. The reactions were run for 40 cycles, and at the end of each run, melt curves were generated to ensure product uniformity. Samples were analyzed in triplicates. Inter-run connector samples were included to correct for the use of multiple plates. All calculations and normalizations were done using the iQ5 software (Bio-Rad).
Vector construction and transformation
The full-length coding sequence of PaHAP3A was amplified from cDNA of proliferating cultures using primers with attB adapters for compliance with the Invitrogen Gateway system (Invitrogen). Full-length coding sequence of PaHAP3A has previously been submitted to GenBank with accession number JF280794 (Uddenberg et al. 2011). All primers are listed in Table S2. To produce an entry clone, the amplified sequence was cloned into the pDONR/Zeo vector, according to the manufacturer’s protocol. All subsequent introductions of the pDONR/Zeo entry clone, containing PaHAP3A, into destination vectors were performed using the Gateway technology according to the manufacturer’s protocol (Invitrogen). An inducible PaHAP3A construct was made by inserting the entry vector into the estradiol-inducible destination vector pMDC7 (Curtis and Grossniklaus 2003). The pMDC7 vector contains the estrogen receptor-based transactivator XVE, which is regulated by the addition of exogenous β-estradiol (Zuo et al. 2000). A constitutive PaHAP3A construct was created by introducing the entry clone into the pMDC32 destination vector. The pMDC32 vector harbors a dual cauliflower mosaic virus (CaMV) 35S promoter for constitutive expression (Curtis and Grossniklaus 2003).
For reporter gene constructs, putative promoter constructs of PaHAP3A were isolated from embryogenic cultures using primers amplifying a 3–4-kb region upstream the ORF based on the scaffold (212005) from the Norway spruce genome 0.8 (www.congenie.org; Nystedt et al. 2013). Primers amplifying the putative promoters contained attB adapters for compliance with the Invitrogen Gateway technology (Invitrogen). Amplified promoter products were cloned into entry clones (pENTR), and PaHAP3A promoter-reporter constructs were created by inserting entry clones into the pGWB3 destination vector (Nakagawa et al. 2007) according to the manufacturer’s protocol (Invitrogen). The pGWB3 vector lack promoter site but contains a N-terminal β-glucuronidase (GUS) sequence after the 3′ recombination site, thus producing the reporter gene construct pPaHAP3A:GUS.
The destination vectors were introduced into the Agrobacterium tumefaciens strain C58:C1, which contains an extra virulence plasmid (pTOK47), through freeze-thawing (Wenck et al. 1999). Cell line 88.22 was used as target for inducible induction and cell line 61.21 for constitutive expression and promoter studies. Embryogenic cultures were transformed by cocultivation with A. tumefaciens according to the protocol of Wenck et al. (1999), with slight modifications (Wenck et al. 1999). Briefly, after initial cocultivation in liquid proliferation medium for 2–7 h, the cultures were collected on filter papers and placed on solidified proliferation medium. After 1 week, the cultures were transferred to proliferation medium supplemented with 10 μg/ml hygromycin, 250 μg/ml cefotaxime, and 250 μg/ml timentin (Ticarcillin and Clavulanate). Thereafter, the cultures were subcultured to fresh culture medium of the same composition every week. Stable transformants were selected after 4 weeks.
Transgenic sublines
Transgenic sublines harboring the GUS gene (35S:GUS, and XVE-GUS), which were used as transformed controls (T-controls), were kindly provided by Alyona Minina (unpublished data). The sublines were derived from cell line 61.21.
Selection of overexpressing transgenic sublines, both constitutive (35S:PaHAP3A) and inducible (XVE-PaHAP3A), for further studies, was done based on transgene expression levels in proliferating PEMs analyzed by qPCR. Three sublines constitutively overexpressing PaHAP3A under the dual strong 35S promoter (35S:PaHAP3A.1, 35S:PaHAP3A.2, and 35S:PaHAP3A.3) (Table S3), and three estradiol-inducible sublines (XVE-PaHAP3A.1, XVE-PaHAP3A.2, and XVE-PAHAP3A.3) (Table S4) showing high transgene expression levels were selected for further analyses.
Induction of the introduced PaHAP3A transcript (XVE-GUS) was effective with both 1 and 10 μM β-estradiol; however, since the expression level in average was five times higher after treatment with 10 μM β-estradiol, this concentration was chosen as standard (data not shown). The induced PaHAP3A expression remained high for at least 10 days (Table S5). Therefore, the cultures were routinely transferred to fresh medium every 10th day.
PaHAP3A transcriptional reporter gene constructs (pPaHAP3A:GUS) were selected based on GUS activity. The transgenic reporter sublines displayed similar staining patterns across multiple tissue samples. Three sublines (pPaHP3A:GUS.1, pPaHAP3A:GUS.2, and pPaHAP3A:GUS.3) were selected for subsequent studies.
Development of mature somatic embryos
The effect of overexpressing PaHAP3A during embryo maturation was assessed by careful examination of embryo progression in three 35S:PaHAP3A sublines (35S:PaHAP3A.1, 35S:PaHAP3A.2, and 35S:PaHAP3A.3) and in the untransformed control. Randomly collected tissue samples from three biological replicates were analyzed weekly, starting from the differentiation of early somatic embryos (−PGR) to 7 weeks on maturation medium. Collected samples were mounted in 1.2 % (w/v) low-melt agarose and assessed for phenotypes and maturation progression under a light microscope (Zeiss Axioplan). The developmental stages of at least 50 embryos per sample were classified as those described by Larsson et al. (2012): stage 1, PEMs; stage 2, early embryos; stage 3, developing late embryos; stage 4, late embryos; and stage 5, early maturing embryos.
Embryogenic potential in germinated somatic embryos
The decrease in embryogenic potential during somatic embryo germination was analyzed in cell line 88.22. Embryos were germinated for 0 to 30 days on germination medium before they were transferred to medium containing PGRs (proliferation medium). The frequency of germinated embryos differentiating new embryogenic tissue as well as the position of the protruding embryogenic tissue was estimated after 5-week incubation on proliferation medium. At least 60 embryos per time point were assessed.
The effect of overexpressing PaHAP3A on the embryogenic potential in germinated somatic embryos was tested in four experimental series:
-
1.
Mature embryos from 37 XVE-PaHAP3A sublines were germinated for 10 days, with or without 10 μM β-estradiol. The germinated embryos were then transferred to medium containing PGRs (proliferation medium). Samples were assessed both for phenotypic deviations during germination and for the ability to stimulate differentiation of embryogenic tissue, which was scored after 5 weeks on proliferation medium. More than 50 germinated embryos per treatment and subline were analyzed.
-
2.
Mature embryos from three XVE-PaHAP3A sublines (XVE-PaHAP3A.1, XVE-PaHAP3A.3, and XVE-aHAP3A.4) were first germinated for 14 days and then subjected to inducible or non-inducible conditions for an additional 5 days on germination medium, before transferred to medium containing PGRs (proliferation medium). The ability to stimulate differentiation of embryogenic tissue was estimated after 5 weeks on proliferation medium. More than 30 germinated embryos per treatment and subline were analyzed.
-
3.
Mature embryos from three 35S:PaHAP3A sublines (35S:PaHAP3A.1, 35S:PaHAP3A.2, and 35S:PaHAP3A.3) and the untransformed control were germinated for 10 days. Thereafter, the germinated embryos were transferred to medium containing PGRs (proliferation medium). The ability to stimulate differentiation of embryogenic tissue was estimated after 5 weeks on proliferation medium. More than 50 germinated embryos per subline were assessed, and the experiment was repeated three times.
-
4.
The effect of ABA treatment on the embryogenic potential was estimated for 10-day-old germinated embryos from XVE-PaHAP3A sublines (XVE-PaHAP3A.1, XVE-PaHAP3A.2, and PaHAP3A.3). The germinated embryos were then treated in liquid germination medium supplemented with (i) 10 μM β-estradiol, (ii) 30 μM ABA, (iii) 10 μM β-estradiol and 30 μM ABA, or (iv) control medium lacking supplements for another 10 days. Thereafter, the germinated embryos were transferred to medium containing PGRs (proliferation medium). The ability to stimulate differentiation of embryogenic tissue was estimated after 5 weeks on proliferation medium. At least 30 germinated embryos per treatment and subline were analyzed
GUS assay
To analyze GUS activity, the samples were first collected in ice-cold 90 % acetic acid and vacuum infiltrated for 10 min at room temperature. Thereafter, the samples were placed on ice and vacuum infiltrated in GUS buffer (50 mM Na2PO4, 0.25 % Triton X-100, 1 mM Fe+III/Fe+IICN, 1 mM Na2EDTA) for 30 min; subsequently, the samples were transferred to fresh GUS buffer supplemented with 1-mM 5-bromo-4-chloro-3-indole β-glucuronic acid for an additional 30 min. The samples were finally released from vacuum and incubated at 37 °C for 2–12 h in darkness.
Histochemical and histological analyses
Mature embryos from subline XVE-PaHAP3A.1 were used to investigate if increased PaHAP3A expression would stimulate accumulation of storage compounds during germination. Mature embryos were germinated on germination medium supplemented with (i) 10 μM β-estradiol, (ii) 30 μM ABA, (iii) 10 μM β-estradiol and 30 μM ABA, or (iv) control medium lacking supplements. After 10 days, the germinating embryos were collected, hand-dissected, and stained for 5 min with Fat Red 7B (Sigma-Aldrich) for visualizing lipid bodies or Lugol (Sigma-Aldrich) for visualizing starch granules. The samples were analyzed in a light microscope (Leica, Leitz DM RX).
To assess the origin of ectopic embryos, mature somatic embryos from XVE-PaHAP3A sublines and the untransformed control were fixed and embedded in histowax (following the embedding method described by Karlgren et al. (2009)). The embryos were serially sectioned at 8 μM using a microtome (Microm, HM 350), and sectioned samples were stained with Toluidine blue O to visualize general anatomical features (Sigma-Aldrich) before analysis in a light microscope (Leica, DMI 4000).
Results and discussion
Expression of PaHAP3A during embryo development
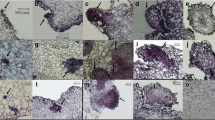
We have previously shown that the expression of PaHAP3A is high during early embryo development; thereafter, the expression decreases to become low in mature embryos and not detectable in 10-day-old germinated embryos (Uddenberg et al. 2011). To gain more insight into the spatial expression of PaHAP3A, histochemical GUS assays were performed with developing and germinated embryos from three selected pPAHAP3A:GUS sublines. In the pPaHAP3A-GUS sublines, strong GUS signals were detected in PEMs (Fig. 1a) and in late embryos (Fig. 1b), both in the embryonal mass and the suspensor. Similar GUS assay results were obtained in PEMs and late embryos from the 35S:GUS subline (Fig. 1f, g). In mature embryos, the GUS signal was stronger in embryos from the 35S:GUS subline (Fig. 1h) than in embryos from the pPaHAP3A:GUS sublines (Fig. 1c). During germination, the GUS signal successively decreased in embryos from the pPaHAP3A:GUS sublines. After 10 days, the GUS staining was localized to an area in the upper hypocotyl surrounding the SAM (Fig. 1d), and sometimes also in the cotyledons, while no GUS signal could be detected in 30-day-old germinated embryos (Fig. 1e). In contrast, the GUS staining was strong throughout the germination in embryos from the 35S:GUS subline (Fig. 1i, j). GUS staining was not detected in embryos from the untransformed control (Fig. 1k–o).
GUS reporter gene expression during embryo development and germination. GUS signal was developed after incubation in GUS solution. Embryos were collected from: a–e a pPaHAP3A:GUS subline, f–j a 35S:GUS subline, and k–o an untransformed control. Developmental stages: a, f, k PEMs, b, g, l late embryos, c, h, m mature embryos, d, i, n embryos germinated for 10 days, and e, j, o embryos germinated for 30 days. Embryos germinated for 30 days were sectioned prior to GUS staining to allow for complete buffer penetrance. MC meristematic cell, VC vacuolated cell, EM embryonal mass, S suspensor, CO cotyledon, HC hypocotyl. Bars at a–c, f–h, k–m 200 μM and d, e, i, j, n, o 2 mm
The GUS assay results are in accordance to our previous results based on qPCR analyses (Uddenberg et al. 2011). However, by using the GUS assay, we could also show a weak PaHAP3A expression in the SAM region in 10-day-old germinated embryos. In angiosperms, LEC1 and its ortholog L1L demonstrate overlapping embryo-specific expression patterns (Lotan et al. 1998; Kwong et al. 2003; Yazawa et al. 2004; Fambrini et al. 2006; Le et al. 2010). Thus, our data show that the expression pattern of PaHAP3A is similar to the expression pattern of LEC1-type genes in angiosperms.
Overexpression of PaHAP3A during embryo maturation stimulates differentiation of ectopic embryos
In order to elucidate if overexpression of PaHAP3A affects embryo maturation and the embryogenic potential, we established transgenic cultures of Norway spruce that overexpressed PaHAP3A, using both constitutive (35S:PaHAP3A sublines) and inducible (XVE-PaHAP3A sublines) promoters (Tables S3 and S4).
The effect of overexpressing PaHAP3A on embryo development was analyzed weekly during the maturation process. The developmental pattern of embryos in the 35S:PaHAP3A sublines was similar to the developmental pattern of embryos in the untransformed control (Fig. 2a), as well as in the XVE-PaHAP3A sublines (data not shown). The endogenous PaHAP3A and PaVP1 genes were expressed in mature embryos both from the PaHAP3A overexpressing sublines and from the control (Fig. S1a, c), while the introduced PaHAP3A gene (transgenic PaHAP3A) was only expressed in mature embryos from the overexpressing sublines (Fig. S1b). These results suggest that overexpression of PaHAP3A does not affect the expression of the endogenous PaHAP3A gene and that the transgenic PaHAP3A gene is expressed during the whole maturation process. Thus, together, our data show that overexpression of PaHAP3A is not affecting the maturation process and that the arrested maturation process caused by TSA treatment (Uddenberg et al. 2011) cannot be explained, at least not solely, by a retained high expression of PaHAP3A. More likely, the retained high expression of PaHAP3A during TSA-treatment is an indirect effect of the continued proliferation of embryogenic tissue and the blocking of the maturation process.
Overexpression of PaHAP3A during embryo maturation. a Proliferating embryogenic tissue from three 35S:PaHAP3A sublines (light gray circles) and the untransformed control (black triangles) were transferred to maturation medium. The developmental stage of the embryos was examined each week from the beginning of maturation treatment (week 0) until 7 weeks on maturation medium. Time points (x-axis) correspond to the number of weeks on maturation medium. Developmental stages (y-axis) are presented as the average developmental stage of more than 50 embryos in each randomly assessed sample and annotated as in Larsson et al. (2012): S1, PEMs; S2, early embryos; S3, developing late embryos; S4, late embryos; S5, early maturing embryos. b–h Mature embryos after 6 weeks on maturation medium. b Mature embryo from the untransformed control. c–h Mature embryo from subline XVE-PaHAP3A.2. The culture had been treated with β-estradiol during the whole maturation process. f–h The same embryos as in c–e, but images are captured from underneath. Images show embryos with differentiating ectopic embryos at different developmental stages: c, f embryogenic tissue (ET), d, g late embryo (LE), e, h early cotyledonary embryo (CE). Ectopic embryos typically protruded from the hypocotyl region subtending the cotyledons. Bars = 200 μm
Interestingly, a subset of the mature embryos from PaHAP3A overexpressing sublines differentiated ectopic embryos from the hypocotyl subtending the cotyledons (Fig. 2c–h). Ectopic embryos were never observed on mature embryos from the controls (more than 1000 mature control embryos were assessed) (Fig. 2b). On average, 5.5 % of the mature embryos from induced XVE-PaHAP3A sublines had differentiated ectopic embryos when transgene expression had been induced from the beginning of the maturation treatment (XVE-PaHAP3A.1 5.4 %, XVE-PaHAP3A.2 5.5 %). In addition, 2.5 % of the non-induced embryos differentiated ectopic embryos (XVE-PaHAP3A.1 2.9 %, XVE-PaHAP3A.2 1.9 %), which most likely is explained by the fact that the XVE promoter is partially activated in Norway spruce even in the absence of β-estradiol (Table S4, Zhu et al. 2014). Ectopic embryos were also observed on mature embryos from 35S:PaHAP3A sublines. The developmental stages of the observed ectopic embryos varied from protruding embryogenic tissue (Fig. 2c, f), to late somatic embryos (Fig. 2d, g), and mature somatic embryos (Fig. 2e, h). Mature embryos with ectopic embryos were sectioned to analyze the origin of the ectopic embryos. Embryos carrying ectopic embryos displayed a wider hypocotyl circumference than control embryos, mainly owing to enlarged cortical cells outside the provascular tissue (Fig. 3c, d). Furthermore, the radial symmetry was lost just below the cotyledons at the position from which the ectopic embryo had differentiated (Fig. 3). This shows that the ectopic embryos had differentiated from the mature embryo explant and not only superficially adhered to the surface.
Transverse sections of a mature somatic embryo displaying a protruding ectopic embryo. a, c, e, g Embryos from the estradiol-induced XVE-PaHAP3A.2 subline display protrusions of ectopic embryos (EE) subtending the base of the cotyledons. b, d, f, h Untransformed control embryos. Dashed lines in g, h indicate the plane of which the radial sections are displayed (a, b upper line, c, d middle line, and e, h lower line). The area inside the red circles in the sections includes provascular tissue and pith, and the area between the yellow and red circles include the inner part of the cortex. a, b Note the aberrant differentiation of cotyledons in the XVE-PaHAP3A embryo (a), indicated by black arrows, compared to the symmetric arrangement of cotyledons in the control embryo (b). c, d Note the enlarged cortical cells and disturbed radial symmetry of the vascular tissue in the XVE-PaHAP3A embryo (c), compared to the control embryo (d). e, f Note that the radial symmetry is similar in e, f. Bars = 100 μm. Transverse sections are stained with Toluidine Blue O
In Arabidopsis, overexpression of LEC1 induces embryonic processes postembryonically even in the absence of plant growth regulators (Lotan et al. 1998; Casson and Lindsey 2006; Junker et al. 2012). Likewise, overexpression of LEC1 in tobacco plants (Nicotiana tabacum) is sufficient to induce embryonic properties in vegetative tissues (Guo et al. 2013). Furthermore, in rare cases, the LEC1 overexpressing phenotype in the Arabidopsis mutants lacking proper LEC1 function has been manifested as differentiation of ectopic embryo-like structures from vegetative tissues (Lotan et al. 1998). Thus, in angiosperms, overexpression of LEC1 can stimulate differentiation of ectopic embryo-like structures. In this work, we show that overexpression of PaHAP3A can stimulate differentiation of ectopic embryos in Norway spruce, which suggests that the conifer LEC1-type gene shares functional similarities with its angiosperm homologue.
We do not know when during the maturation process, the ectopic embryos on mature somatic embryos of Norway spruce are initiated, but they differentiate just below the cotyledons and seem to originate from cells neighboring provascular tissues. It has been shown that pericycle cells in the hypocotyl of both angiosperms (reviewed in Verstraeten et al. 2014) and gymnosperms (Grönroos and von Arnold 1988) have the potential to differentiate adventitious roots. In Arabidopsis, stimulation of calli from various tissues, including aerial postembryonic tissues, leads to ectopic activation of a pericycle reporter, and the proliferating callus cells differentiate and express genes with characteristics of the root meristem (Sugimoto et al. 2010). It is therefore possible that similar processes occur in gymnosperms, although the establishment of reporter genes for different cell fates is required before comparisons and firm conclusions can be made. Recent advances in single cell omics and the release of the Norway spruce genome (Nystedt et al. 2013; Junker and van Oudenaarden 2014) may now supply possibilities to further investigate the origin of the ectopic embryos.
Overexpression of PaHAP3A is not sufficient to increase the embryogenic potential in germinated embryos
It has previously been shown that the frequency of germinated conifer embryos differentiating embryogenic tissue decreases successively as the age of the germinated embryos increases (Klimaszewska et al. 2010; Uddenberg et al. 2011). In Norway spruce, the decrease of the embryogenic potential includes both a decrease in the frequency of germinated embryos that initiate embryogenic tissue and a decrease in the number of sites on the embryo explants from which embryogenic tissue protrude (Fig. S2). To test whether overexpression of PaHAP3A affects the embryogenic potential, germinated embryos from 35S:PaHAP3A and XVE-PaHAP3A sublines were transferred to proliferation medium to stimulate differentiation of embryogenic tissue. Overexpression of PaHAP3A did not affect the germination process (data not shown). No significant change in the embryogenic potential could be observed between germinated embryos from transgenic sublines overexpressing PaHAP3A and controls, (Supplementary Note). Our results that overexpression of PaHAP3A is not sufficient to postembryonically stimulate an increase of the embryogenic potential are in line with previous reports. In angiosperms, it has been shown that overexpression of AtLEC1 in tobacco seedlings is not sufficient to stimulate differentiation of embryogenic tissue (Guo et al. 2013). Accordingly, it was not possible to stimulate differentiation of embryogenic tissue in germinated somatic embryos of white spruce (Picea glauca) derived from a transgenic cell line overexpressing a HAP3A gene (Klimaszewska et al. 2010). Based on the results from Klimaszewska et al. (2010) and observations of the expression levels of a HAP3A gene in Pinus radiata callus lines with varying degrees of embryogenic potential, Garcia-Mendiguren et al. (2015) suggested that high expression levels of HAP3A alone is not enough for establishing embryogenic cultures and that additional embryonic regulators are likely required to confer embryogenic potential in conifers (Garcia-Mendiguren et al. 2015). In Arabidopsis, Junker et al. (2012) showed that overexpression of LEC1 alone causes minor effects on seedlings germinated for 10 days, followed by 10 days of induced LEC1 expression. However, overexpression of LEC1 together with addition of exogenous ABA causes seedlings to inhibit vegetative development and adopt maturation characteristics, such as an accumulation of seed storage compounds (Junker et al. 2012). Overexpression of PaHAP3A in the presence of ABA did not affect the embryogenic potential in germinated embryos of Norway spruce (data not shown), neither could we observe any obvious effect on the accumulation of fatty acids and starch (Fig. S3). Thus, our results suggest that overexpression of PaHAP3A, with or without exogenous ABA, does not cause any postembryonic maturation properties in germinated Norway spruce embryos, in contrast to what has been shown for LEC1-type genes in angiosperms.
The pattern of embryogenic tissue differentiation changes when the embryos germinate (Fig. S2). When mature zygotic embryos of Norway spruce were used as explants, usually, embryogenic tissue was protruding from the whole embryo (Fig. S2b). In contrast, when germinated embryos were used as explants, embryogenic tissue differentiated from more localized regions (Fig. S2c–e). Before the primary explants are overgrown by calli, it is evident that embryogenic tissue preferentially differentiates from the upper part of the hypocotyl or from the cotyledons (Uddenberg et al. 2011). Simultaneously, as the embryogenic potential decreased, the formation of non-embryogenic callus increased (Fig. S2d, e, and f). To elucidate a putative colocalization of the expression of PaHAP3A and the differentiation of embryogenic tissue, germinated embryos from pPaHAP3A:GUS sublines were transferred to proliferation medium to stimulate initiation of embryogenic tissue. Explants were assessed for GUS expression patterns after 4 weeks on proliferation medium, a time point corresponding to just before embryogenic tissue could be visualized. The expression of PaHAP3A, according to GUS assay results, was mainly localized to the upper part of embryos germinated for 10 days (Fig. 4a, d). Patchy patterns of GUS signals were detected along the cotyledons. In embryos germinated for 30 days, only a weak diffuse GUS staining was observed with occasional GUS foci in the upper hypocotyl bordering the cotyledons (Fig. 4g, j). In contrast, strong GUS staining was detected in germinated embryos from the 35S:GUS subline (Fig. 4b, e, h, k), while no GUS staining was observed in germinated embryos from the untransformed control (Fig. 4c, f, i, l). The patchy expression pattern of PaHAP3A in the cotyledons and upper hypocotyl of germinated pPaHAP3A:GUS embryos, before the embryogenic tissue protrude, coincide with the place at which embryogenic tissue normally differentiate. It is thus tempting to speculate that PaHAP3A may be important for the embryogenic potential, and although it might not by itself be enough to stimulate embryogenic tissue formation, it may very well be an early genetic maker of tissues harboring embryogenic potential.
Expression pattern of PaHAP3A in germinated embryos according to GUS reporter assays. Germinated embryos were generated from a, d, g, j a pPaHAP3A:GUS subline, b, e, h, k a 35S:GUS subline, and c, f, i, l an untransformed control. a–c Embryos were germinated for 10 days or g–i 30 days, before transferred to proliferation medium to stimulate differentiation of embryogenic tissue. Images are captured after 4 weeks on proliferation medium, which is just before the embryogenic tissue protrude from the explant. Rectangles in a–c and g–i indicate approximate areas of images in d–f and j–l, respectively. a, d, g, j Note that the fragmentary GUS signals in germinated embryos from the pPaHAP3A:GUS subline mainly localize to the upper part of the explant, whereas in b, e, h, k, the GUS signals are more evenly distributed in the germinated embryos from 35S:GUS subline. No GUS expression was detected in the control embryos (c, f, i, l). Bars = 1 mm
Conclusions
In this study, we show that PaHAP3A is expressed at presumptive sites of new embryogenic tissue, suggesting that the gene might be an early genetic maker of tissues harboring embryogenic potential. However, in contrast to what has been shown for other LEC1-type genes in angiosperms, overexpression of PaHAP3A is not sufficient to promote embryonic properties postembryonically. Overexpression of PaHAP3A during the maturation stage does not affect the maturation process, but a subset of the mature embryos differentiate ectopic embryos. The ectopic embryos most likely differentiate from cells neighboring provascular tissues just below the cotyledons, which indicates that the hormonal and or cellular status in these cells may be required to augment the effect of PaHAP3A overexpression. Future studies using novel gene identification methods, e.g., cell/tissue-specific transcriptome analyses, will become useful to unravel the molecular processes underlying the capacity of somatic cells to differentiate somatic embryos.
References
Alvarez JM, Sohlberg J, Engström P et al (2015) The WUSCHEL-RELATED HOMEOBOX 3 gene PaWOX3 regulates lateral organ formation in Norway spruce. New Phytol 208:1078–1088. doi:10.1111/nph.13536
Boutilier K, Offringa R, Sharma VK et al (2002) Ectopic expression of BABY BOOM triggers a conversion from vegetative to embryonic growth. Plant Cell 14:1737–1749. doi:10.1105/tpc.001941
Braybrook S, Harada J (2008) LECs go crazy in embryo development. Trends Plant Sci 13:624–630. doi:10.1016/j.tplants.2008.09.008
Casson SA, Lindsey K (2006) The turnip mutant of Arabidopsis reveals that LEAFY COTYLEDON1 expression mediates the effects of auxin and sugars to promote embryonic cell identity. Plant Physiol 142:526–541. doi:10.1104/pp.106.080895
Curtis MD, Grossniklaus U (2003) A gateway cloning vector set for high-throughput functional analysis of genes in planta. Plant Physiol 133:462–469. doi:10.1104/pp.103.027979
Fambrini M, Durante C, Cionini G et al (2006) Characterization of LEAFY COTYLEDON1-LIKE gene in Helianthus annuus and its relationship with zygotic and somatic embryogenesis. Dev Genes Evol 216:253–264. doi:10.1007/s00427-005-0050-7
Gaj MD, Zhang S, Harada JJ, Lemaux PG (2005) Leafy cotyledon genes are essential for induction of somatic embryogenesis of Arabidopsis. Planta 222:977–988. doi:10.1007/s00425-005-0041-y
Garcia-Mendiguren O, Montalbán IA, Stewart D et al (2015) Gene expression profiling of shoot-derived calli from adult radiata pine and zygotic embryo-derived embryonal masses. PLoS ONE 10, e0128679. doi:10.1371/journal.pone.0128679
Goldberg RB, de Paiva G, Yadegari R (1994) Plant embryogenesis: zygote to seed. Science 266:605–614. doi:10.1126/science.266.5185.605
Grönroos R, von Arnold S (1988) Initiation of roots on hypocotyl cuttings of Pinus sylvestris, with emphasis on direct rooting, root elongation, and auxin uptake. Can J For Res 18:1457–1462
Guo F, Liu C, Xia H et al (2013) Induced expression of AtLEC1 and AtLEC2 differentially promotes somatic embryogenesis in transgenic tobacco plants. PLoS ONE 8, e71714. doi:10.1371/journal.pone.0071714
Gutierrez L, Van Wuytswinkel O, Castelain M, Bellini C (2007) Combined networks regulating seed maturation. Trends Plant Sci 12:294–300. doi:10.1016/j.tplants.2007.06.003
Harding EW, Tang W, Nichols KW et al (2003) Expression and maintenance of embryogenic potential is enhanced through constitutive expression of AGAMOUS-Like 15. Plant Physiol 133:653–663. doi:10.1104/pp.103.023499
Hecht V, Vielle-Calzada JP, Hartog MV et al (2001) The Arabidopsis SOMATIC EMBRYOGENESIS RECEPTOR KINASE 1 gene is expressed in developing ovules and embryos and enhances embryogenic competence in culture. Plant Physiol 127:803–816
Junker JP, van Oudenaarden A (2014) Every cell is special: genome-wide studies add a new dimension to single-cell biology. Cell 157:8–11. doi:10.1016/j.cell.2014.02.010
Junker A, Mönke G, Rutten T et al (2012) Elongation-related functions of LEAFY COTYLEDON1 during the development of Arabidopsis thaliana. Plant J 71:427–442. doi:10.1111/j.1365-313X.2012.04999.x
Karlgren A, Carlsson J, Gyllenstrand N et al (2009) Non-radioactive in situ hybridization protocol applicable for Norway spruce and a range of plant species. J Vis Exp. doi:10.3791/1205
Klimaszewska K, Pelletier G, Overton C et al (2010) Hormonally regulated overexpression of Arabidopsis WUS and conifer LEC1 (CHAP3A) in transgenic white spruce: implications for somatic embryo development and somatic seedling growth. Plant Cell Rep 29:723–734. doi:10.1007/s00299-010-0859-z
Kwong R, Bui A, Lee H et al (2003) LEAFY COTYLEDON1-LIKE defines a class of regulators essential for embryo development. Plant Cell 15:5–18. doi:10.1105/tpc.006973
Larsson E, Sundström JF, Sitbon F, von Arnold S (2012) Expression of PaNAC01, a Picea abies CUP-SHAPED COTYLEDON orthologue, is regulated by polar auxin transport and associated with differentiation of the shoot apical meristem and formation of separated cotyledons. Ann Bot 110:923–934. doi:10.1093/aob/mcs151
Le BH, Cheng C, Bui AQ et al (2010) Inaugural article: global analysis of gene activity during Arabidopsis seed development and identification of seed-specific transcription factors. Proc Natl Acad Sci 107:8063–8070. doi:10.1073/pnas.1003530107
Lotan T, Ohto M, Yee K et al (1998) Arabidopsis LEAFY COTYLEDON1 is sufficient to induce embryo development in vegetative cells. Cell 93:1195–1205
Nakagawa T, Kurose T, Hino T et al (2007) Development of series of gateway binary vectors, pGWBs, for realizing efficient construction of fusion genes for plant transformation. J Biosci Bioeng 104:34–41. doi:10.1263/jbb.104.34
Nystedt B, Street NR, Wetterbom A et al (2013) The Norway spruce genome sequence and conifer genome evolution. Nature 497:579–584. doi:10.1038/nature12211
Ogas J, Cheng JC, Sung ZR, Somerville C (1997) Cellular differentiation regulated by gibberellin in the Arabidopsis thaliana pickle mutant. Science 277:91–94
Ogas J, Kaufmann S, Henderson J, Somerville C (1999) PICKLE is a CHD3 chromatin-remodeling factor that regulates the transition from embryonic to vegetative development in Arabidopsis. Proc Natl Acad Sci U S A 96:13839–13844
Rider S, Henderson J, Jerome R et al (2003) Coordinate repression of regulators of embryonic identity by PICKLE during germination in Arabidopsis. Plant J 35:33–43. doi:10.1046/j.1365-313X.2003.01783.x
Schmidt ED, Guzzo F, Toonen MA, de Vries SC (1997) A leucine-rich repeat containing receptor-like kinase marks somatic plant cells competent to form embryos. Development 124:2049–2062
Sugimoto K, Jiao Y, Meyerowitz EM (2010) Arabidopsis regeneration from multiple tissues occurs via a root development pathway. Dev Cell 18:463–471. doi:10.1016/j.devcel.2010.02.004
Tai HH, Tai GCC, Beardmore T (2005) Dynamic histone acetylation of late embryonic genes during seed germination. Plant Mol Biol 59:909–925. doi:10.1007/s11103-005-2081-x
Tanaka M, Kikuchi A, Kamada H (2007) The Arabidopsis histone deacetylases HDA6 and HDA19 contribute to the repression of embryonic properties after germination. Plant Physiol 146:149–161. doi:10.1104/pp.107.111674
ten Hove CA, Lu K-J, Weijers D (2015) Building a plant: cell fate specification in the early Arabidopsis embryo. Development 142:420–430. doi:10.1242/dev.111500
Uddenberg D, Valladares S, Abrahamsson M et al (2011) Embryogenic potential and expression of embryogenesis-related genes in conifers are affected by treatment with a histone deacetylase inhibitor. Planta 234:527–539. doi:10.1007/s00425-011-1418-8
Vandesompele J, De Preter K, Pattyn F, et al. (2002) Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol 3:–. doi: 10.1186/gb-2002-3-7-research0034
Verstraeten I, Schotte S, Geelen D (2014) Hypocotyl adventitious root organogenesis differs from lateral root development. Front Plant Sci. doi:10.3389/fpls.2014.00495
Vestman D, Larsson E, Uddenberg D et al (2011) Important processes during differentiation and early development of somatic embryos of Norway spruce as revealed by changes in global gene expression. Tree Genet Genomes 7:347–362. doi:10.1007/s11295-010-0336-4
von Arnold S, Clapham D (2008) Spruce embryogenesis. Methods in Molecular Biology. Humana Press, Totowa, NJ. 427:31–47
Wenck AR, Quinn M, Whetten RW et al (1999) High-efficiency Agrobacterium-mediated transformation of Norway spruce (Picea abies) and loblolly pine (Pinus taeda). Plant Mol Biol 39:407–416. doi:10.1023/A:1006126609534
West M (1993) Embryogenesis in higher plants: an overview. Plant Cell Oneline 5:1361–1369. doi:10.1105/tpc.5.10.1361
Yazawa K, Takahata K, Kamada H (2004) Isolation of the gene encoding Carrot leafy cotyledon1 and expression analysis during somatic and zygotic embryogenesis. Plant Physiol Biochem 42:215–223. doi:10.1016/j.plaphy.2003.12.003
Zhu T, Moschou PN, Alvarez JM, et al (2014) Wuschel-related homeobox 8/9 is important for proper embryo patterning in the gymnosperm Norway spruce. J Exp Bot 65:6543–6552. doi:10.1093/jxb/eru371
Zuo J, Niu QW, Chua NH (2000) Technical advance: an estrogen receptor-based transactivator XVE mediates highly inducible gene expression in transgenic plants. Plant J 24:265–273
Zuo J, Niu QW, Frugis G, Chua NH (2002) The WUSCHEL gene promotes vegetative-to-embryonic transition in Arabidopsis. Plant J 30:349–359
Acknowledgments
The authors would like to thank Alyona Minina for GUS control constructs and Iva Mozgova for critical reading of the manuscript. This work was supported by the Swedish Research Council Formas.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Data archiving statement
The full-length CDS of PaHAP3A (accession number JF280794) has been submitted to GenBank.
Additional information
Communicated by A. Brunner
Electronic supplementary material
Below is the link to the electronic supplementary material.
ESM 1
(PDF 2050 kb)
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Uddenberg, D., Abrahamsson, M. & von Arnold, S. Overexpression of PaHAP3A stimulates differentiation of ectopic embryos from maturing somatic embryos of Norway spruce. Tree Genetics & Genomes 12, 18 (2016). https://doi.org/10.1007/s11295-016-0974-2
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11295-016-0974-2