Abstract
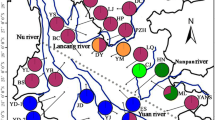
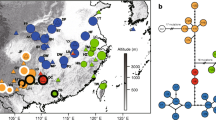
The Amentotaxus argotaenia (Taxaceae) species complex is comprised of four relict and endangered gymnosperms. Amentotaxus argotaenia and A. yunnanensis are scattered throughout South and Central China, and A. formosana, and A. poilanei are restricted to Taiwan and Vietnam. In the present study, we examined genetic divergence and biogeographical patterns in the species complex based on chloroplast DNA atpI-atpH and rpS16-trnK intergenic spacers, the mitochondrial DNA nad1 intron, and microsatellite markers. Genealogical analyses revealed systematic inconsistencies between organellar DNA markers, with paraphyletic species inferred based on cpDNA, versus a monophyletic A. formosana, and clustering of A. poilanei within A. yunnanensis at mtDNA. AMOVA revealed that most organellar DNA variants resided among species and populations, a pattern also supported by clustering with STRUCTURE analyses on microsatellites. Higher levels of genetic diversity in the Yongxin population of A. argotaenia reflected features of a refugium. Most speciation events were rather recent, with divergence no earlier than 2.50 MYA, although many ancient lineages derived 4.12–13.84 MYA were maintained within species. Significant genetic structuring within A. argotaenia was detected among three geographical regions—a phylogeographical pattern likely attributable to past fragmentations. In contrast to paraphyletic A. argotaenia based on the maintenance of ancestral polymorphisms, interspecific hybridization likely contributed to polyphyly in A. yunnanensis. Furthermore, despite the low likelihood of recurrent gene flow due to geographical isolation, IM analyses revealed continued interspecific gene flow after species divergence, reflecting a parapatric speciation model.





Similar content being viewed by others
References
Axelrod I, Ai-Shehba I, Raven PH (1996) History of the modern flora of China. In: Zhang A, Wu A (eds) Floristic characteristics and diversity of East Asian plants. Springer, New York, pp 43–55
Belshaw R, Katzourakis A (2005) BlastAlign: a program that uses blast to align problematic nucleotide sequences. Bioinformatics 21:122–123
Cheng WC (1978) Flora Republicae Popularis Sinicae. vol. 7, The genus Amentotaxus, vol 7. Science, Beijing
Chiang TY, Schaal BA (2006) Phylogeography of plants in Taiwan and the Ryukyu Archipelago. Taxon 55:3–41
Chiang TY, Chiang YC, Chou CH, Havanond S, Hong TN, Huang S (2001) Phylogeography of Kandelia candel in East Asiatic mangroves based on nucleotide variation of chloroplast and mitochondria DNAs. Mol Ecol 10:2697–2710
Chiang TY, Hung KH, Hsu TW, Wu WL (2004) Lineage sorting and phylogeography in Lithocarpus formosanus and L. dodonaeifolius (Fagaceae) from Taiwan. Ann Mo Bot Gard 91:207–222
Chiang YC, Hung KH, Schaal BA, Ge XJ, Hsu TW, Chiang TY (2006) Contrasting phylogeographical patterns between mainland and island taxa of the Pinus luchensis complex. Mol Ecol 15:765–779
Chiang YC, Hung KH, Moore SJ, Ge XJ, Huang S, Hsu TW, Schaal BA, Chiang TY (2009) Paraphyly of organelle DNAs in Cycas sect. Asiorientales due to ancient ancestral polymorphisms. BMC Evol Biol 9:161
Comes HP, Kaderaeit JW (1998) The effect of Quaternary climatic changes on plant distribution and evolution. Trends Plant Sci 3:432–438
DeChaine EG, Martin AP (2005) Marked genetic divergence among sky island populations of Sedum lanceolatum (Crassulaceae) in the Rocky Mountains. Am J Bot 92:477–486
Del Tredici P, Lin G, Yang G (1992) The Ginkgos of Tian Mu Shan. Conserv Biol 4:202–209
Dias P, Assis LCS, Udulutsch RG (2005) Monophyly vs paraphyly in plant systematics. Taxon 54:1039–1040
Doyle JJ, Doyle JL (1987) A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull 19:11–15
Drummond AJ, Ho SY, Phillips MJ, Rambaut A (2006) Relaxed phylogenetics and dating with confidence. PLoS Biol 4:e88
Earl DA, vonHoldt BM (2012) STRUCTURE HARVESTER: a website and program for visualizing STRUCTURE output and implementing the Evanno method. Conserv Genet Resour 4:359–361
Evanno G, Regnaut S, Goudet J (2005) Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Mol Ecol 14:2611–2620
Excoffier L, Smouse PE (1994) Using allele frequencies and geographic subdivision to reconstruct gene trees within a species: molecular variance parsimony. Genetics 136:343–359
Excoffier L, Smouse PE, Quattro JM (1992) Analysis of molecular variance inferred from metric distances among DNA haplotypes: application to human mitochondrial DNA data. Genetics 131:479–491
Excoffier L, Laval G, Schneider S (2005) Arlequin (version 3.0): an integrated software package for population genetics data analysis. Evol Bioinform Online 1:47–50
Farjon A (2010) A handbook of the world’s conifers. Brill, Leiden
Felsenstein J (1985) Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39:783–791
Felsenstein J (2007) PHYLIP (Phylogeny Inference Package), version 3.67, Department of Genetics, University of Washington, Seattle.
Ferguson DK, Jahnichen H, Alvin KL (1978) Amentotaxus Pilger from the European Tertiary. Feddes Repert 89:379–410
Fu YX (1997) Statistical tests of neutrality of mutations against population growth, hitchhiking, and background selection. Genetics 147:915–925
Fu YX, Li WH (1993) Statistical tests of neutrality of mutations. Genetics 133:693–709
Ge XJ, Zhou XL, Li ZC, Hsu TW, Schaal BA, Chiang TY (2005) Genetic structure of the relict Amentotaxus argotaenia complex (Taxaceae) based on ISSR fingerprinting. J Plant Res 118:415–422
Ge XJ, Hsu TW, Hung KH, Lin CJ, Huang CC, Huang CC, Chiang YC, Chiang TY (2012) Inferring multiple refugia and phylogeographical patterns in Pinus massoniana based on nucleotide sequence variation and fingerprinting. PLoS One 7:e43717
Goudet J (1995) FSTAT Version 2.9.3.2: a computer program to calculate F-statistics. J Hered 86:485–486
Groth JG, Barrowclough GF (1999) Basal divergences in birds and the phylogenetic utility of the nuclear RAG-1 gene. Mol Phylogenetd Evol 12:115–123
Harris TM (1976) Two neglected aspects of fossil conifers. Am J Bot 63:902–910
Harrison SP, Yu G, Takahar H, Prentice IC (2001) Diversity of temperate plants in east Asia. Nature 413:129–130
Hewitt GM (1996) Some genetic consequences of ice ages, and their role in divergence and speciation. Biol J Linn Soc 58:247–276
Hewitt GM (1999) Post-glacial re-colonization of European biota. Biol J Linn Soc 68:87–112
Hewitt GM (2000) The genetic legacy of the Quaternary ice ages. Nature 405:907–913
Hey J, Nielsen R (2004) Multilocus methods for estimating population sizes, migration rates, and divergence time, with applications to the divergence of Drosophila pseudoobscura and D. persimilis. Genetics 167:747–760
Ho CS, Chen CT, Ko YZ, Ge XJ, Hung KH, Hsu TW, Chiang YC (2012) Isolation and characterization of 15 microsatellite loci in four endangered Amentotaxus species (Taxaceae). Am J Bot 99:e157–159
IUCN (2013) The IUCN Red List of Threatened Species. Version 2013.2 http://www.iucnredlist.org
Kalinowski ST (2005) HP-RARE 1.0: a computer program for performing rarefaction on measures of allelic richness. Mol Ecol Notes 5:187–189
Leslie AB, Beaulieu JM, Rai HS, Crane PR, Donoghue MJ, Mathews S (2012) Hemisphere-scale differences in conifer evolutionary dynamics. Proc Natl Acad Sci USA 109:16217–16221
Li HL, Keng H (1994) Amentotaxaceae. In: Editorial Committee of the Flora of Taiwan (ed) Flora of Taiwan, 2nd edn. Vol. 1. Department of Botany, National Taiwan University, Taipei, pp 553–554
Lin CC (1966) An outline of Taiwan’s Quaternary geohistory with a special discussion of the relation between natural history and cultural history in Taiwan. Bull Dept Archaeol Anthropol 23:7–44
Liu KB (1988) Quaternary history of the temperate forests of China. Q Sci Rev 7:1–20
Liu HP (1991) The spore-pollen communities and paleoenvironment since Quaternary in the reach of Chang Jiang River. J Huazhong Normal Univ Nat Sci 2:236–241
Matos JA, Schaal BA (2000) Chloroplast evolution in the Pinus montezumae complex: a coalescent approach to hybridization. Evolution 54:1218–1233
Melo-Ferreira J, Boursot P, Carneiro M, Esteves PJ, Farelo L, Alves PC (2012) Recurrent introgression of mitochondrial DNA among hares (Lepus spp.) revealed by species-tree inference and coalescent simulations. Syst Biol 61:367–381
Moriguchi Y, Kang KS, Lee KY, Lee SW, Kim YY (2009) Genetic variation of Picea jezoensis populations in South Korea revealed by chloroplast, mitochondrial and nuclear DNA markers. J Plant Res 122:153–160
Müller K (2005) SeqState - primer design and sequence statistics for phylogenetic DNA data sets. Appl Bioinform 4:65–69
Nei M (1987) Molecular evolutionary genetics. Columbia University Press, New York
O’Connell LM, Ritland K (2004) Somatic mutations at microsatellite loci in western red cedar (Thuja plicata: Cupressaceae). J Hered 95:172–176
Parchman TL, Benkman CW, Jenkins B, Buerkle CA (2011) Low levels of population genetic structure in Pinus contorta (Pinaceae) across a geographic mosaic of co-evolution. Am J Bot 98:669–679
Peng Y, Yin S, Wang J, Tian B, Ren G, Guo Q, Liu J (2012) Phylogeographic analysis of the fir species in southern China suggests complex origin and genetic admixture. Ann Forest Sci 69:409–416
Petit RJ, Aguinagalde I, de Beaulieu JL, Bittkau C, Brewer S, Cheddadi R, Ennos R, Fineschi S, Grivet D, Lascoux M, Mohanty A, Müller-Starck G, Demesure-Musch B, Palmé A, Martín JP, Rendell S, Vendramin GG (2003) Glacial refugia: hotspots but not melting pots of genetic diversity. Science 300:1563–1565
Pons O, Petit RJ (1996) Measuring and testing genetic differentiation with ordered versus unordered alleles. Genetics 144:1237–1245
Potter D, Freudenstein JV (2005) Character-based phylogenetic Linnaean classification: taxa should be both ranked and monophyletic. Taxon 54:1033–1035
Pritchard J, Wen W (2003) Documentation for STRUCTURE Software, version 3.2. http://pritch.bsd.uchicago.edu/structure.html
Pritchard J, Stephens M, Donnelly P (2000) Inference of population structure using multilocus genotype data. Genetics 155:945–959
Rambaut A, Drummond A (2004) Tracer–MCMC trace analysis tool. University of Oxford, Oxford
Ray N, Adams JM (2001) A GIS-based vegetation map of the world at the last glacial maximum (25,000-15,000 BP). Internet Archaeology 11. Available from: http://www.ncdc.noaa.gov/paleo/pubs/ray2001/ray_adam_2001.pdf
Rogers AR, Harpending H (1992) Population growth makes waves in the distribution of pairwise genetic differences. Mol Biol Evol 9:552–569
Royer DL, Hickey LJ, Wing SL (2003) Ecological conservatism in the “living fossil” Ginkgo. Paleobiology 29:84–104
Rozas J, Sánchez-DelBarrio JC, Messeguer X, Rozas R (2003) DnaSP, DNA polymorphism analyses by the coalescent and other methods. Bioinformatics 19:2496–2497
Slatkin M, Hudson RR (1991) Pairwise comparisons of mitochondrial DNA sequences in stable and exponentially growing populations. Genetics 129:555–562
Stewart WN, Rothwell GW (1993) Paleobotany and the evolution of plants, 2nd edn. Cambridge University Press, Cambridge
Taberlet P, Fumagalli L, Wust-Saucy AG (1998) Comparative phylogeography and postglacial colonization routes in Europe. Mol Ecol 7:453–464
Tajima F (1989) Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics 123:585–595
Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S (2011) MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28:2731–2739
Tong G, Zhang J, Fan S (1992) Distribution of Quaternary palynoflora in China. Marine Geol Quat Geol 12:45–56
Tong GB, Chen Y, Wu XH, Li ZH, Yang ZJ, Wang SB, Cao JD (1999) Pleistocene environmental megaevolution and indicated by the sporopollen floras in China. J Geophys Res 5:11–21
Tran TTT (2011) Spatial distribution and historical dynamics of threatened conifers of the Dalat Plateau. Master thesis. University of Missouri, Columbia
Traula H (1968) The phytogeographic evolution of the genus Ginkgo L. Bot Notiser 120:409–422
Vendramin GG, Anzidei M, Madaghiele A, Sperisen C, Bucci G (2000) Chloroplast microsatellite analysis reveals the presence of population subdivision in Norway spruce (Picea abies K.). Genome 43:68–78
Wachowiak W, Prus-Glowacki W (2008) Hybridisation processes in sympatric populations of pines Pinus sylvestris L., P. mugo Turra and P. uliginosa Neumann. Plant Syst Evol 271:29–40
Wang JP, Hsu KC, Chiang TY (2000) Mitochondrial DNA phylogeography of Acrossocheilus paradoxus (Cyprinidae) in Taiwan. Mol Ecol 9:1483–1494
Wang JF, Pan YZ, Gong X, Chiang YC, Kuroda C (2011) Chloroplast DNA variation and phylogeography of Ligularia tongolensis (Asteraceae), a species endemic to the Hengduan Mountains region of China. J Syst Evol 49:108–119
Wang J, Abbott RJ, Peng YL, Du FK, Liu JQ (2012) Species delimitation and biogeography of two fir species (Abies) in central China: cytoplasmic DNA variation. Heredity 107:362–370
Wang JF, Gong X, Chiang YC, Kuroda C (2013) Phylogenetic patterns and disjunct distribution in Ligularia hodgsonii Hook. (Asteraceae). J Biogeogr 40:1741–1754
Watterson GA (1975) On the number of segregating sites in genetical models without recombination. Theor Popul Biol 7:256–276
Wiens JJ, Penkrot TA (2002) Delimiting species using DNA and morphological variation and discordant species limits in spiny lizards (Sceloporus). Syst Biol 51:69–91
Wolfe KH, Li WH, Sharp PM (1987) Rates of nucleotide substitution vary greatly among plant mitochondrial, chloroplast, and nuclear DNAs. Proc Natl Acad Sci USA 84:9054–9058
Xu JX, Ferguson DK, Li CS, Wang YF (2008) Late Miocene vegetation and climate of the Lühe region in Yunnan, southwestern China. Rev Palaeobot Palynol 48:36–59
Zhou QX (2001) Studies on systematics of the Taxaceae. Dissertation, Kunming Institute of Botany, Kunming, China
Zhou B, Li G, Yao M, Fang L (1994) An analysis on the periglacial geomorphy and paleogeographic environment during the Quaternary of the Mount Jiuhuashan. J Anhui Normal Univ Natur Sci 17:48–53
Zhou YF, Abbott RJ, Jiang ZY, Du FK, Milne RI, Liu JQ (2010) Gene flow and species delimitation: a case study of two pine species with overlapping distributions in southeast China. Evolution 64:2342–2352
Zou J, Sun Y, Li L, Wang G, Yue W, Lu Z, Wang Q, Liu J (2013) Population genetic evidence for speciation pattern and gene flow between Picea wilsonii, P. morrisonicola and P. neoveitchii. Ann Bot 112:1829–1844
Acknowledgments
We thank Chao-Yi Deng, Guo-Sheng He, Yun-Fei Deng and Hua-Gu Ye for their help with material sampling. We also thank Joanna Schultz (Lewis-Clark State College, ID) and two reviewers for their helpful comments. This study was supported financially by National Basic Research Program of China (973 Program) (Grant No. 2014CB954100) National Natural Science Foundation of China (No. 30170070), and the National Science Council of Taiwan (NSC 96-2628-B-020 -001 -MY2 and NSC 102-2628-B-110 -001 -MY3).
Author information
Authors and Affiliations
Corresponding authors
Additional information
X.-J. Ge and K.-H. Hung contributed equally to this work.
Electronic supplementary material
Below is the link to the electronic supplementary material.
ESM 1
(DOC 77 kb)
Rights and permissions
About this article
Cite this article
Ge, XJ., Hung, KH., Ko, YZ. et al. Genetic Divergence and Biogeographical Patterns in Amentotaxus argotaenia Species Complex. Plant Mol Biol Rep 33, 264–280 (2015). https://doi.org/10.1007/s11105-014-0742-0
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11105-014-0742-0