Abstract
Background and Aims
Lespedeza cuneata (Dum. Cours.) G. Don is an invasive legume that displaces populations of native N. American congeners. Our aims are to determine the growth benefits of different rhizobacterial strains for L. cuneata and native Lespedeza virginica (L.) Britton, and to determine if these strains influence competition between these plants.
Methods
Plants were grown under nitrogen-limiting conditions in sterilized soil in pairs consisting of two L. cuneata, two L. virginica, or one of each species, and then plants were inoculated with one of seven rhizobial isolates, or with a no-strain control. After 3 months, plants were harvested for determination of biomass and nodulation rate.
Results
Five of the assayed stains improved L. cuneata biomass over uninoculated controls, but none of the strains benefited L. virginica. L. cuneata plants had more biomass and root nodules when grown in competition with L. virginica than with a conspecific.
Conclusions
Asymmetrical benefits from these symbionts accrued to invasive L. cuneata but not to native L. virginica, and this may provide the invader with a growth advantage in the field. Changes in the availability of effective symbionts in the soils of invaded sites can shape performance of native and invasive plants.
Similar content being viewed by others
Introduction
Lespedeza cuneata (commonly known as Sericea lespedeza) is a long-lived perennial legume in the family Fabaceae and also one of the notorious invasive plants of North America. This plant was introduced from Japan to the U.S. in the 1800s, and since then it has become an invasive weed, causing ecological problems in its introduced range (Eddy and Moore 1998). L. cuneata can tolerate high drought and shade conditions and survive in various habitats including prairies, woodlands, fields and borders of ponds and swamps (Remaley 1998). It is known for its capability of causing changes in plant community composition and the structure and function of native habitats (Brandon et al. 2004; Eddy and Moore 1998). The symbiotic relationship between L. cuneata and nitrogen-fixing bacteria has the potential to alter soil nutrients in the ecosystem (Hawkes et al. 2005; Lynd and Ansman 1993a). These soil nutrient changes can lead to vegetation or microbial community alteration (Hawkes et al. 2005). Due to its aggressive behavior and its harmful effects, L. cuneata has been put on the noxious weed list in several states (Dudley and Fick 2003), which calls for more insightful understanding of the invasion of L. cuneata.
Several possible ecological mechanisms of invasion success of L. cuneata have been studied by previous workers. L. cuneata is more productive than its North American congeners, having a higher total and specific leaf area, and this productivity allows L. cuneata to outcompete native species (Allred et al. 2010; Smith and Knapp 2001). L. cuneata can tolerate a large amount of leaf loss (80 %) during the growing season while maintaining a similar growth rate to unclipped controls (Schutzenhofer and Knight 2007). A study conducted by Beaton et al. (2011) found evidence that L. cuneata from its introduced sites is more competitive than native plants and its ancestral genotype. What is more, invasive genotypes of L. cuneata exhibit enhanced aggressiveness (Beaton et al. 2011) in accordance with the evolution of increased competitive ability hypothesis. This hypothesis predicts that a lack of natural enemies allows invasive plants to allocate more resources to growth and fewer resources to defense in their invaded range, and thus they tend to grow more biomass and be more effective light competitors than individuals of the same plant species taken from its original native sites (Blossey and Notzold 1995). These characteristics build the foundation of the “shade-out” effect of L. cuneata, and there is evidence that L. cuneata tends to lower the light availability, allowing it to dominate grassland communities under favorable conditions (Brandon et al. 2004).
In addition to its growth advantage, L. cuneata may interact with its environment in ways that confer benefit over native competitors. L. cuneata can consistently maintain significant levels of both seed and vegetative reproduction under variable weather conditions, facilitating the spread of this species under a wide range of environmental conditions, including those that might otherwise affect flowering, pollination, seed dispersal, germination and establishment (Woods et al. 2009). Furthermore, the release of phytotoxic compounds from plant residues of L. cuneata has been shown to inhibit several crops and weeds (Kalburtji et al. 2001).
Interaction with the soil microbial community provides another, little explored mechanism of the invasive success of L. cuneata. An increasing number of studies indicate that interactions between plants and microorganisms can influence plant invasions (Klironomos 2002; Mitchell et al. 2006; Reinhart and Callaway 2006; van der Putten 2010; van der Putten et al. 2007; Wolfe and Klironomos 2005), and mutualisms with rhizobacteria should be particularly important for the success of invasive legumes like L. cuneata. Many leguminous plants continue to fix nitrogen after invading a new habitat, by taking advantage of mutualisms with bacteria that were transported with them, or by forming new mutualisms in the invaded range (De Faria et al. 1989). Consequently, some invasive legumes do not appear to be limited by the density or community composition of soil rhizobia (Birnbaum et al. 2012). However, the legume-rhizobium symbiosis is more effective when invasive Acacia are matched with bacteria from their home range in comparison with native bacteria (Rodriguez-Echeverria et al. 2012; Thrall et al. 2007). The legume Leucaena leucocephala failed to survive in its invaded range until its corresponding symbiotic rhizobia were introduced (Richardson et al. 2000). What is more, a threshold density of nitrogen fixing bacteria is required for nodule forming on some legumes (Parker 2001), and some invasive legumes may be limited by the distribution of their symbionts (Parker et al. 2006). Thus, the symbiotic relationship with rhizobia still has important functions for plant growth after invading a new habitat.
Although it is known that obtaining nitrogen from root nodules is important for invasive legumes in habitats with low levels of nitrogen, it is still not known how the mutualism with rhizobia may help invasive legumes outcompete native vegetation. There are different mechanisms that nodule formation might influence plant competition, thereby impacting plant invasion. One of the possible mechanisms is that certain legumes may be superior hosts because they are relatively more adept at acquiring symbiotic partners than other legumes (Rodríguez-Echeverría et al. 2009). Also, the effectiveness of rhizobia varies dramatically from host to host indicating that certain plants will be favored if a site has more rhizobacteria that are more beneficial for these plants as opposed to potential competitors (Kiers et al. 2003).
While it has come to be recognized that soil microorganisms associated with plants (e.g. rhizobacteria and mycorrhizal fungi (Reinhart and Callaway 2006)) might play an important role in invasion success (Klironomos 2002; Mitchell et al. 2006; Reinhart and Callaway 2006; van der Putten et al. 2007), few studies have reported how the interaction between L. cuneata and its associated microorganisms influences its invasion success. However, a recent study (Yannarell et al. 2011) about L. cuneata suggests that soil bacteria communities in heavily invaded and uninvaded sites were significantly different, which raises the possibility that microorganisms associated with L. cuneata invasion may play a role in facilitating its invasion.
Mechanisms by which nodule-forming bacteria might influence plant competition may be operating in the spread of the invasive L. cuneata at the expense of its native congener, L. virginica (Schutzenhofer and Knight 2007). Both legumes readily associate with N-fixing Rhizobiales bacteria, but they vary widely in their densities where range overlap occurs. This might be due to their physiological and morphological differences, but we hypothesize that it is also related to their different responses to symbiosis with rhizobia under identical biotic and abiotic conditions. By forming nodules with L. cuneata and L. virginica, different rhizobial strains might benefit L. cuneata and L. virginica with different efficiency.
Here we ask, how do different rhizobial strains influence the competition between L. cuneata and its native congener, L. virginica under low-nitrogen conditions? We tested the effects of cultivated rhizobial strains on both invasive L. cuneata and native L. virginica when grown separately and in competition. Our objectives are to determine whether these rhizobia provide asymmetrical benefits to L. cuneata and L. virginica, and to evaluate the potential impacts of these symbioses on competition between these two plant species.
Materials and methods
Isolation and screening of bacterial strains
We collected roots and root nodules of invasive Lespedeza cuneata (Dum. Cours.) G. Don, native L. virginica (L.) Britton and a native legume, Chamaechrista fasciculata (Michx.) Greene var. fasciculata from previously described locations (Yannarell et al. 2011) at Ft. Benning, Georgia and Ft. Leonard Wood, Missouri. Root nodules were cleaned with 0.02 % polysorbate 20, surface sterilized with 6 % sodium hypochlorite and then rinsed with autoclaved DI water three times (Bender et al. 1989). Root nodules were crushed into 500 μl autoclaved DI water. Aliquots of 100 μl of this bacteria liquid were spread gently over Yeast-Manitol agar medium plates, and then after 3 to 7 days of growth at 28 °C, single colonies were picked and transferred to R2A medium plates for isolation. Colonies were continually picked and transferred to R2A plates until one single colony type was seen on the plate. We obtained 50 isolates in this manner. Each single colony was collected from isolated culture plates to enrich on a new R2A medium plate. After several days of growth at 28 °C, isolated bacteria were swabbed into 50 % glycerol stock for long-term storage.
The remaining bacteria liquid on the swab of each culture was suspended in autoclaved DI water for identification by 16S rDNA gene sequencing. The bacterial suspension was boiled in 100 °C water bath for 10 min to release DNA, and then 5 μl of this liquid was used as a template for 16S rRNA gene PCR. The 16S rRNA gene PCR was done using the primers of Lane (Lane 1991) based on the following conditions: each 50 μl reaction contained 5 μl template, GoTaq (Promega, Madison, WI) buffer 1×, 0.25 μg μl−1 BSA, 3 mM MgCl2, 0.25 μM dNTPs, primer 8 F (5’-GGGTTBCCCCATTCRG-3’) 0.4 μM, primer 1492R (5’-GGGTTBCCCCATTCRG-3’) 0.4 μM, Taq polymerase (Promega, Madison, WI) 0.05U/μl; program: 94 °C 2 min; 30 cycles of 94 °C 35 s, 55 °C 45 s, 72 °C 2 min; 72 °C 2 min. The PCR products were purified using a Promega PCR cleanup kit and sent to the W.M. Keck Center for Comparative and Functional Genomics (University of Illinois at Urbana-Champaign) for Sanger sequencing (Sanger and Coulson 1975) for each strain. The strains were then identified by 16S rRNA gene sequences (Yanagi and Yamasato 1993). All sequences generated as part of this work have been deposited in GenBank under accession numbers KF596683-KF596726.
Isolates were further screened for the presence of nitrogenase genes (nifH) and a functional nitrogenase system. PCR for the nifH gene was conducted from the boiled bacteria DNA liquid, using the primer set of Poly and colleagues (Poly et al. 2001) based on the following conditions: each 25 μl reaction contained 4 μl template, GoTaq buffer (Promega, Madison, WI) 1×, 0.25 μg μl−1 bovine serum albumin (BSA), 2 mM MgCl2, 0.2 μM dNTPs, primer polF (5’-TGCGAYCCSAARGCBGACTC-3’) 0.5 μM, primer polR (5’-ATSGCCATCATYTCRCCGGA-3’) 0.5 μM, Taq enzyme (Promega, Madison, WI) 0.05 U μl−1; program: 94 °C 5 min; 1 cycle of 94 °C 45 s, 64 °C 45 s, 72 °C 45 s, 2 cycles of 94 °C 45 s, 62 °C 45 s, 72 °C 45 s, 3 cycles of 94 °C 45 s, 60 °C 45 s, 72 °C 45 s, 4 cycles of 94 °C 45 s, 58 °C 45 s, 72 °C 45 s, 25 cycles of 94 °C 45 s, 56 °C 45 s, 72 °C 45 s, 72 °C 10 min. Gene products of the expected size were confirmed by electrophoresis. To confirm that strains could actively fix nitrogen, we assayed all nifH-positive strains for growth on nitrogen-free AcD plates (Burris 1994). We continued checking the plates to see if the bacteria strains grew on the plates for as long as 2 weeks.
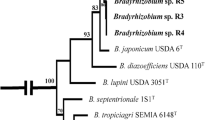
Thirteen isolated strains belonging to traditional nodule-forming bacterial genera were able to grow on the nitrogen-free medium. Based on comparisons of the 16S rRNA gene sequences, we found that several of our isolates were 100 % identical to others. Thus, we narrowed isolates down to seven unique rhizobial strains (Table 1) used to test our hypotheses regarding bacterial influence on plant competition.
Competition trials
A greenhouse experiment was conducted to find out how different rhizobial strains influence the competition between L. cuneata and L. virginica. The experiment was a three-way factorial experiment to test the influence of rhizobial strain identity on plant competition. The plant factor had two levels, L. cuneata and L. virginica. The competition factor was assessed by growing two L. cuneata individuals alone (intraspecific competition), two L. virginica individuals alone (intraspecific competition), and one individual of the two species together (interspecific competition). Seven different rhizobial strains were used to inoculate these plants, and a no-inoculum control was also used (8 levels total). Ten replicates were applied to each treatment.
We used a standard “root wash” soil mixture provided by the University of Illinois’ Plant Care Facility greenhouse. This soil mixture consisted of 1:1:1 of field soil: calcinated clay: sand), and it had the following properties (mean ± standard deviation): organic matter (1.65 % ± 0.90 %), total organic carbon (0.95 % ± 0.10 %), total nitrogen (0.079 % ± 0.002 %), Bray-1 phosphorus (25.5 ppm ± 6.56 ppm), nitrate (7.5 ppm ± 1.0 ppm), ammonium (8.0 ppm ± 0.8 ppm), pH (7.3 ± 0.05), cation exchange capacity (14.2 meq/100 g ± 1.1 meq/100 g). This soil mixture was autoclaved twice at 121 °C for 1 h. Approximately 450 cubic centimeters of soil was put into separated Leonard jars (Leonard 1943). These growth containers consisted of two 77 mm × 77 mm × 97 mm (W × L × H) polycarbonate pots (GA-7 Magenta vessels, Sigma Aldrich, St. Louis, MO), one on top of the other, connected by a wick. The lower box contained water and nutrients for plants, which were carried by the wick up to the upper box that contains soil and plants. All Leonard jars with soil were autoclaved for 25 min with lids on.
Half-strength sterilized nitrogen-free Hoagland’s solution (Bender et al. 1989) was diluted from autoclaved stock, and 200 ml was added to the lower portion of each autoclaved Leonard jar. The soil in the upper compartment was moistened by nutrient solution for 2 days before dispersing seeds on the soil.
All seeds were sterilized with 6 % sodium hypochlorite for 5 min, and rinsed with sterile Milli Q water three times (Bender et al. 1989). Eight seeds were put in each Leonard jar with moistened soil. Between the 9th and 11th day after germination, two seedlings were randomly selected and left in the jar as experimental subjects while others were culled. The seedlings were inoculated with strains 7–14 days after germination. The seven rhizobial strains were grown to the end of exponential stage in Yeast-Manitol broth at 28 °C, with shaking at 160 rpm. The strains were then diluted to 108 CFU/ml for inoculation, and every seedling was inoculated with 5 ml bacteria suspension liquid.
During germination, lids with a small central vent covered with a 0.2 μm filter were put on Leonard jars to prevent contamination in the beginning phase. The lids were kept on after inoculation for 5–7 days to prevent cross contamination from the air. Fresh, sterile half-strength nitrogen-free Hoagland’s solution was added to the bottom compartment of the Leonard jars each week in order to replenish all nutrients except for nitrogen. At the end of experiment, the plants were large enough that the frequency of Hoagland’s solution resupply was increased to two to three times a week. The pots were randomized to different locations on the greenhouse bench top weekly.
After 3 months of growth, plants were harvested. Both plants, along with the attached soil, were removed from the Leonard jars. The two plants from the same pot were then gently separated from each other and the soil. Plants were washed with tap water to remove soil, and roots and shoots were separated from washed plants. Root nodule number was counted before each shoot and root was separated and dried for biomass measurement. Nodules were removed from roots and weighed on a three-digit scale to help explain the performance variation between plant individuals. The rest of roots were put back into the envelope. All envelopes with shoots and roots were dried at 60 °C. After 3 days, the biomass of shoot and root was measured on a three-digit scale.
Data analysis
To evaluate differences in root biomass, shoot biomass, total plant biomass, nodule number, and nodule biomass, three-way ANOVAs were conducted in the R statistical environment (v2.14.1, R Development Core Team, 2011). As described above, the factors for these analyses were plant species (two levels), competition (two levels, inter- and intraspecific competition), and bacterial strain (eight levels, including un-inoculated controls). Post-hoc contrasts using Tukey’s Honest Significant Difference were conducted to identify significant differences between specific treatment levels.
Results
The main effects of plant species, competition, and bacterial strain were significant for root biomass, shoot biomass, total plant biomass, nodule biomass, and nodule number (Tables 2, S1–3). L. cuneata acquired more biomass (Figs. 1 and 2) and more nodules (Fig. 3) than L. virginica. Plants were larger under interspecific competition than intraspecific competition, but this effect seemed to be largely driven by the superior performance of L. cuneata under interspecific competition (Fig. 2). The two-way interaction of plant species and strain were significant in total biomass (Table 2) and shoot biomass (Table S1), meaning that the growth of L. cuneata and L. virginica reacted differently to the strain treatments. L. cuneata gained significantly more biomass than the no-inoculation control when inoculated with R2 (Bradyrhizobium1) (p < 0.001, Fig. 1), R3 (Rhizobium1) (p = 0.011, Fig. 1), R4 (Bradyrhizobium2) (p < 0.001, Fig. 1), R5 (Rhizobium2) (p = 0.013, Fig. 1) and R7 (Bradyrhizobium3) (p = 0.014, Fig. 1). However, none of the strains yielded significant increases in L. virginica biomass in comparison to the no-strain control (p = 0.5541, Fig. 1).
Influence of rhizobial strain identity on plant biomass The data show the total biomass of L. cuneata (black) and L. virginica (white) plants when inoculated with no strain (leftmost set of bars) or with one of the seven strains listed in Table 1 (strain identities are as follows: R1 Mesorhizobium; R2 Bradyrhizobium1; R3 Rhizobium1; R4 Bradyrhizobium2; R5 Rhizobium2; R6 Rhizobium3; R7 Bradyrhizobium3). Error bars represent one standard error of the mean
Influence of plant competition on plant biomass The data show the total biomass of L. cuneata (black) and L. virginica (white) plants when grown under intraspecific competition (e.g. L. cuneata plant with L. cuneata neighbor; leftmost black bar) or under interspecific competition (e.g. L. cuneata plant with L. virginica neighbor, black bar on the right). Error bars represent one standard error of the mean
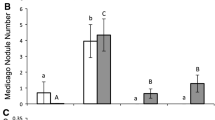
Influence of plant competition on plant nodulation rate The data show the number of root nodules for L. cuneata (black) and L. virginica (white) plants when grown under intraspecific competition (e.g. L. cuneata plant with L. cuneata neighbor; leftmost black bar) or under interspecific competition (e.g. L. cuneata plant with L. virginica neighbor, black bar on the right). Error bars represent one standard error of the mean
In addition, the two-way interaction of plant species and competition were significant for total biomass (Table 2) and shoot biomass (Table S1), meaning that the growth of L. cuneata and L. virginica reacted differently to the competition treatment. For L. cuneata, the total biomass (p < 0.001, Fig. 2) and the nodule number (p < 0.001, Fig. 2) were significantly larger under interspecific competition than under intraspecific competition, whereas there was no such difference for L. virginica. Overall L. cuneata gained significantly more total biomass (Tables 2 and 4), shoot biomass (Table 4, S1), root biomass (Table 4, S2), nodule biomass (Table 4, S3), and nodule number (Tables 3 and 4) than L. virginica. L. cuneata also had a higher root-to-shoot biomass ratio than L. virginica (p < 0.001), but there were no other significant treatment effects or interactions for root-to-shoot biomass.
Discussion
Asymmetrical benefit of microbial mutualists to native and invasive plants
Our results demonstrate that native and invasive Lespedeza do not benefit equally from associations with their rhizobial symbionts. Plant biomass of L. cuneata inoculated with R2 (Bradyrhizobium1), R3 (Rhizobium1), R4 (Bradyrhizobium2), R5 (Rhizobium2) and R7 (Bradyrhizobium3) was significantly higher than uninoculated controls, whereas the plant biomass of L. virginica did not improve over that of controls with any of the strains tested here (Fig. 1). This asymmetric benefit of particular strains for L. cuneata led to improvements in total biomass and shoot biomass and to trend-level improvements in nodule number (Tables 2 and 3, S1). No such benefits were observed for L. virginica, even when coupled with strains that were originally isolated from L. virginica plants (e.g. strain R6). L. cuneata acquired more nodule biomass than L. virginica (Table 4), which may mean that the invasive L. cuneata was able to establish a more profitable nitrogen symbiosis than the native. Regardless of the mechanism, it is clear that invasive L. cuneata is disproportionately benefiting from some of the strains surveyed here, while the native L. virginica is not. L. cuneata has previously been reported to associate with mycorrhizal fungi (Lynd and Ansman 1993b; Wilson 1988), and while we are aware of no reports on the mycorrhizal status of L. virginica, a number of other Lespedezas are reported to benefit from arbuscular mycorrhizal fungi (Maki et al. 2008; Wu et al. 2002). It would be very interesting to know if the disproportionate benefits of L. cuneata reported here extend to these other kinds of symbioses.
Different strains of microbial symbionts can have differential fitness effects on different plant hosts (Cardinale et al. 2008; Helgason et al. 2002; Rangin et al. 2008; Rincon-Rosales et al. 2009; Rodriguez-Echeverria et al. 2012; van der Heijden et al. 1998), and these differential effects may be related to the degree of “familiarity” between the host and the symbiont (Klironomos 2003; Rodriguez-Echeverria 2010; Rodriguez-Echeverria et al. 2012). For example, in a Mediterranean dune system, the legume-rhizobium symbiosis leads to higher nodulation rates, nitrogenase activity, and plant growth when native legumes are paired with native rhizobacteria and when exotic legumes are paired with rhizobacteria from their home range (Rodriguez-Echeverria et al. 2012; Thrall et al. 2007). These differential effects can link the dynamics of plant and soil communities (Bever et al. 2012). As plant community composition changes, the dominant plants can influence the diversity of soil mutualists through their symbiont preference (Leary et al. 2006; Rangin et al. 2008). A plant experiencing a declining population of its preferred symbionts may be colonized by a higher proportion of inferior strains, and the resulting mismatch between plant hosts and effective symbionts may have detrimental effects on plant fitness. If the legume-rhizobium symbiosis of native plants is less effective with strains that are preferred by invasive plants, then dominance of invasive plants at a site can exert additional pressure on native populations through a kind of “invasional meltdown” of their preferred symbionts (Rodriguez-Echeverria 2010; Rodriguez-Echeverria et al. 2012). Our results do not allow us to make any conclusions about plant preference for symbionts, but we can point to differences in the benefits obtained by the invader and the native (Fig. 1). Furthermore, our previous work has shown that high densities of invasive L. cuneata are correlated with shifts in overall soil bacterial community composition in comparison to uninvaded sites (Yannarell et al. 2011). Further research is necessary before it can be established that these belowground changes can reinforce L. cuneata invasion, but our current results indicate that changes to the composition of nitrogen-fixing rhizobacteria in invaded habitats could lead to asymmetrical benefits for L. cuneata over its native congeners.
Understanding how shifts in microbial community composition can influence invasion success will help build our ecological toolkit for managing plant invasions, and it is particularly important to understand which species might play a key role in supporting plant invasion. The three Bradyrhizobium strains tested here were all effective nodulators of invasive L. cuneata, and these three strains also yielded the highest total biomass increase for this plant (Fig. 1). As with the other strains tested here, none of these Bradyrhizobium strains produced positive biomass increases for the native L. virginica. In addition, one of these effective Bradyrhizobium strains was isolated from a root nodule of the native plant Chamaechrista fasciculata (strain R7), and this raises the potential for facilitative interactions between C. fasciculata and L. cuneata by means of their common symbionts, as has been previously demonstrated for invasive Cystisus scoparius and native Desmodium canadense (Parker et al. 2006). Previous studies have implicated Bradyrhizobium strains in supporting exotic legume invasions in New Zealand (Weir et al. 2004), Australia (Lafay and Burdon 2006), Africa (Boukhatem et al. 2012; Ndlovu et al. 2013), Europe (Rodriguez-Echeverria 2010; Rodríguez-Echeverría et al. 2007; Rodriguez-Echeverria et al. 2012), and Hawai’i (Leary et al. 2006). Weir and colleagues report that Bradyrhizobium was the only genus associated with three different invasive legumes in New Zealand (Weir et al. 2004), and Rodríguez-Echevarría and colleagues similarly report that all isolates from different invasion stages of Acacia longifolia were in the genus Bradyrhizobium (Rodríguez-Echeverría et al. 2007). The ability of invasive Acacia to associate with Bradyrhizobium may be responsible for its successful invasion of Algeria, where the native Acacia associate exclusively with Rhizobium and Mesorhizobium (Boukhatem et al. 2012). Finally, there are a number of reports of co-invasion by exotic legumes and their associated Bradyrhizobium bacteria (Lafay and Burdon 2006; Leary et al. 2006; Rodriguez-Echeverria 2010; Rodriguez-Echeverria et al. 2012). In conjunction with the widespread reports of Bradyrhizobium associations with plant invasions, the large asymmetry Bradyrhizobium-associated plant growth for invasive and native Lespedeza reported here makes Bradyrhizobium a likely candidate genus for further study in regards to L. cuneata invasion.
Our work demonstrates that mutualist strain identity can influence plant performance, but we acknowledge that our selection of seven strains only scratches the surface of potential plant-microbe interactions of relevance to L. cuneata invasion. By screening for strains capable of growth on nitrogen-free plates (i.e. able to fix nitrogen in the free-living state), we excluded strains that only fix nitrogen as mutualists in root nodules, and these organisms may have important ecological consequences for native and invasive Lespedezas. In addition, 16S rRNA gene sequencing revealed that some of our initial root nodule isolates belonged to genera that are not traditionally considered to be root nodule mutualists (i.e. Burkholderia). Determining whether these organisms have direct influence on Lespedeza competition or whether they influence the performance of nodule-forming rhizobacteria should further enhance our understanding of microbial roles in L. cuneata invasion.
Plant competition
The outcome of plant competition can depend both on the relative densities of the competing plant species and on the total density of plants. For this reason, plant competition experiments may follow substitutive designs (constant total plant density with different ratios of competitor species), additive designs (constant density of a focal plant species with different densities of neighboring plants), or response surface designs (densities of both competitor species are varied) (Goldberg and Scheiner 1993). Because our main goal was to evaluate the potential impact of multiple bacterial strains on competition between Lespedeza plants, we used a relatively simple substitutive design to maintain a manageable number of experimental units. However, our use of a substitutive design means that we were only able to assess the relative intensity of intra- and interspecific competition (Scheiner and Gurevitch 1993) in the presence and absence of our bacterial strains. Translation of our results to field-relevant predictions of L. cuneata invasions requires a more detailed accounting of density-dependent effects of L. cuneata and L. virginica, such as could be obtained with a response surface experimental design. Given our results on the variable impact of different rhizobial genera, future work could pursue such response surface designs using native and invasive Lespedezas and their symbionts, for example, by focusing on one of the key Bradyrhizobium strains. Regardless of these limitations, our data on the relative strength of intra- and interspecific competition between Lespedezas suggests some interesting ecological patterns.
We found significant plant, competitor, and plant x competitor interactions for total biomass, shoot biomass, and root biomass (Table 2, S1–2). This is because invasive L. cuneata was consistently larger (Table 4) than native L. virginica in all trials, and it acquired much more biomass (root and shoot) when grown in competition with L. virginica than when grown under intraspecific competition (Fig. 2). In contrast, L. virginica showed no difference in total biomass in competition with L. cuneata as compared to competition with itself. These results are consistent with previous studies regarding the competitive advantages of L. cuneata (Allred et al. 2010; Brandon et al. 2004; Richardson et al. 2000). We also sought to determine if the presence or identity of rhizobial symbionts could influence the competition between these two plants. However, we found no significant competitor x strain (or plant x competitor x strain) interactions on plant biomass (Table 2, S1–3), and thus we found no evidence that these strains differentially influence the competition between L. cuneata or L. virginica.
Association with nitrogen-fixing bacteria can influence the competition between legumes and non-legumes. Legumes can increase the amount of symbiont-derived nitrogen when growing with non-nitrogen-fixing plants (Hagan and Jose 2011; Karpenstein-Machan and Stuelpnagel 2000), which can decrease the overall strength of competition for nitrogen through niche partitioning. However, Bauer and colleagues have proposed the increased competition with legumes was responsible for the decrease in grass abundance in greenhouse mesocosms to which nitrogen-fixing rhizobacteria had been added (Bauer et al. 2012). It is likely that the influence of nitrogen-fixing rhizobacteria on competition between legumes and non-legumes depends largely on whether the limiting resource is nitrogen (making niche partitioning a possibility) or something else (e.g. sunlight). Similar considerations should apply to legume-legume competition. Thus, under conditions of light or water limitation, we would expect that L. cuneata would have a competitive edge over L. virginica due to its larger shoot and root biomass. Under nitrogen-limiting conditions, such as those present in our experiment, legumes should be able to rely on their symbionts, provided that effective symbionts are present.
In this regard, our data provide some interesting patterns. As discussed above, L. cuneata was much more likely to benefit from the symbiosis than was L. virginica, which may mean that the native L. virginica experienced nitrogen-limiting conditions throughout our trials. Also, L. cuneata experienced significantly higher nodulation rates when grown in competition with L. virginica than when grown under intraspecific competition (Fig. 3). This may indicate that the L. cuneata plants had to compete with each other over effective rhizobacterial symbionts under intraspecific competition. We note that there was a trend level competitor x strain interaction for nodulation rate (Table 3), which is consistent with the notion that the identity of neighboring plants (i.e. inter- versus intraspecific competition) can affect the ability of some symbionts to form nodules on Lespedeza plants.
Although the continual supply of nitrogen-free Hoagland’s solution throughout the growth period was intended to keep our plants under nitrogen-limited conditions, we cannot rule out the possibility that plant growth was also limited by other factors (space, water, light, nutrients). Thus, while it is possible that increased nodulation of L. cuneata under interspecific competition led to higher growth rates for these plants, it is also possible that the increased nodulation rate of L. cuneata was a result of its faster growth rate. In competition with slower growing native L. virginica, the larger L. cuneata may be superior at acquiring resources (nutrients, water, or light) from the limited soil volume in our experimental pots, which could stimulate nodulation of L. cuneata. Both of these explanations are consistent with our observations, and this highlights the need to understand the primary limiting factors for plant growth when studying plant-microbe mutualisms.
Conclusions
Here we have shown that invasive Lespedeza cuneata was able to benefit more often from rhizobacterial mutualists than its native congener, L. virginica. This differential benefit means that the identity and availability of rhizobacterial symbionts in the soil can influence plant performance, such that any changes that diminish the availability of preferred symbionts of the native plant can have negative consequences for native plant growth at a site. Furthermore, because invasive L. cuneata derived a growth benefit from a Bradyrhizobium strain isolated from another native legume (C. fasciculata), it is possible that these plants can facilitate each other by means of their mutual symbionts. While we found no evidence that rhizobacterial strain identity affected the magnitude or direction of competitive outcomes for these plants, we found that invasive L. cuneata had higher overall nodulation rates when grown in combination with L. virginica than when grown with a conspecific. Thus, plant community composition can have an important effect on the successful establishment of the legume-rhizobium symbiosis. Soil organisms and plants are tied together in ecological feedback loops by the influence of plant communities on the diversity and activity of soil borne mutualists and by the asymmetrical influences of different mutualist strains on plant performance.
Abbreviations
- PCR:
-
Polymerase chain reaction
References
Allred BW, Fuhlendorf SD, Monaco TA, Will RE (2010) Morphological and physiological traits in the success of the invasive plant Lespedeza cuneata. Biol Invasions 12:739–749
Bauer JT, Kleczewski NM, Bever JD, Clay K, Reynolds HL (2012) Nitrogen-fixing bacteria, arbuscular mycorrhizal fungi, and the productivity and structure of prairie grassland communities. Oecologia 170:1089–1098. doi:10.1007/s00442-012-2363-3
Beaton LL, Van Zandt PA, Esselman EJ, Knight TM (2011) Comparison of the herbivore defense and competitive ability of ancestral and modern genotypes of an invasive plant, Lespedeza cuneata. Oikos
Bender DA, Neal JL, Morse RD (1989) Evaluation of rhizobial strains nodulating lespedeza cuneata for improving N input into minesoil revegetation systems 1. Commun in Soil Sci & Plant Anal 20:1033–1044
Bever JD, Platt TG, Morton ER (2012) Microbial population and community dynamics on plant roots and their feedbacks on plant communities. In: S Gottesman, CS Harwood, O Schneewind (eds) Annual Review of Microbiology, Vol 66
Birnbaum C, Barrett LG, Thrall PH, Leishman MR (2012) Mutualisms are not constraining cross-continental invasion success of Acacia species within Australia. Divers Distrib 18:962–976. doi:10.1111/j.1472-4642.2012.00920.x
Blossey B, Notzold R (1995) Evolution of increased competitive ability in invasive nonindigenous plants: a hypothesis. J Ecol 83:887–889
Boukhatem ZF, Domergue O, Bekki A, Merabet C, Sekkour S, Bouazza F, Duponnois R, Lajudie P, Galiana A (2012) Symbiotic characterization and diversity of rhizobia associated with native and introduced acacias in arid and semi-arid regions in Algeria. FEMS Microbiology Ecology
Brandon AL, Gibson DJ, Middleton BA (2004) Mechanisms for dominance in an early successional old field by the invasive non-native Lespedeza cuneata (Dum. Cours.) G. Don. Biol Invasions 6:483–493
Burris R (1994) Comparative study of the response of Azotobacter vinelandii and Acetobacter diazotrophicus to changes in pH. Protoplasma 183:62–66
Cardinale M, Lanza A, Bonni ML, Marsala S, Puglia AM, Quatrini P (2008) Diversity of rhizobia nodulating wild shrubs of Sicily and some neighbouring islands. Arch Microbiol 190:461–470. doi:10.1007/s00203-008-0394-2
De Faria S, Lewis G, Sprent J, Sutherland J (1989) Occurrence of nodulation in the Leguminosae. New Phytologist: 607–619
Dudley DM, Fick WH (2003) Effects of sericea lespedeza residues on selected tallgrass prairie grasses. Trans Kans Acad Sci 106:166–170
Eddy T, Moore C (1998) Effects of sericea lespedeza (Lespedeza cuneata (Dumont) G. Don) invasion on oak savannas in Kansas. Trans Wis Acad Sci Arts Lett 86:57–62
Goldberg DE, Scheiner SM (1993) ANOVA and ANCOVA: Field competition experiments. In: SM Scheiner, J Gurevitch (eds) Design and Analysis of Ecological Experiments. Chapman & Hall, New York, NY
Hagan DL, Jose S (2011) Interspecific competition enhances nitrogen fixation in an actinorhizal shrub. Plant Ecol 212:63–68. doi:10.1007/S11258-010-9803-0
Hawkes CV, Wren IF, Herman DJ, Firestone MK (2005) Plant invasion alters nitrogen cycling by modifying the soil nitrifying community. Ecol Lett 8:976–985
Helgason T, Merryweather JW, Denison J, Wilson P, Young JPW, Fitter AH (2002) Selectivity and functional diversity in arbuscular mycorrhizas of co-occurring fungi and plants from a temperate deciduous woodland. J Ecol 90:371–384. doi:10.1046/J.1365-2745.2001.00674.X
Kalburtji K, Mosjidis J, Mamolos A (2001) Allelopathic plants 2. Lespedeza cuneata. Allelopathy J 8:41–50
Karpenstein-Machan M, Stuelpnagel R (2000) Biomass yield and nitrogen fixation of legumes monocropped and intercropped with rye and rotation effects on a subsequent maize crop. Plant Soil 218:215–232. doi:10.1023/A:1014932004926
Kiers ET, Rousseau RA, West SA, Denison RF (2003) Host sanctions and the legume-rhizobium mutualism. Nature 425:78–81
Klironomos JN (2002) Feedback with soil biota contributes to plant rarity and invasiveness in communities. Nature 417:67–70
Klironomos JN (2003) Variation in plant response to native and exotic arbuscular mycorrhizal fungi. Ecology 84:2292–2301
Lafay B, Burdon JJ (2006) Molecular diversity of rhizobia nodulating the invasive legume Cytisus scoparius in Australia. J Appl Microbiol 100:1228–1238. doi:10.1111/j.1365-2672.2006.02902.x
Lane DJ (1991) 16S/23S rRNA sequencing. In: E Stackebrandt, M Goodfellow (eds) Nucleic Acid Techniques in Bacterial Systematics. Wiley, New York
Leary JK, Hue NV, Singleton PW, Borthakur D (2006) The major features of an infestation by the invasive weed legume gorse (Ulex europaeus) on volcanic soils in Hawaii. Biol Fert Soils 42:215–223. doi:10.1007/s00374-005-0018-9
Leonard LT (1943) A simple assembly for use in the testing of cultures of rhizobia. J Bacteriol 45:523
Lynd J, Ansman T (1993a) Symbiotic tripartite nitrogen fixation effectual in eroded soil restoration with 20 year age Sericea lespedeza. J Plant Nutr 16:149–164
Lynd JQ, Ansman TR (1993b) Symbiotic tripartite nitrogen fixation effectual in eroded soil restoration with 20 year-age Sericea lespedeza. J Plant Nutr 16:149–164
Maki T, Nomachi M, Yoshida S, Ezawa T (2008) Plant symbiotic microorganisms in acid sulfate soil: significance in the growth of pioneer plants. Plant Soil 310:55–65. doi:10.1007/S11104-008-9628-Y
Mitchell CE, Agrawal AA, Bever JD, Gilbert GS, Hufbauer RA, Klironomos JN, Maron JL, Morris WF, Parker IM, Power AG (2006) Biotic interactions and plant invasions. Ecol Lett 9:726–740
Ndlovu J, Richardson DM, Wilson JRU, Le Roux JJ (2013) Co-invasion of South African ecosystems by an Australian legume and its rhizobial symbionts. J Biogeogr 40:1240–1251. doi:10.1111/jbi.12091
Parker MA (2001) Mutualism as a constraint on invasion success for legumes and rhizobia. Divers Distrib 7:125–136
Parker MA, Malek W, Parker IM (2006) Growth of an invasive legume is symbiont limited in newly occupied habitats. Divers Distrib 12:563–571. doi:10.1111/j.1366-9516.2006.00255.x
Poly F, Monrozier LJ, Bally R (2001) Improvement in the RFLP procedure for studying the diversity of nifH genes in communities of nitrogen fixers in soil. Res Microbiol 152:95–103. doi:10.1016/S0923-2508(00)01172-4
Rangin C, Brunel B, Cleyet-Marel JC, Perrineau MM, Bena G (2008) Effects of Medicago truncatula genetic diversity, rhizobial competition, and strain effectiveness on the diversity of a natural Sinorhizobium species community. Appl Environ Microbiol 74:5653–5661. doi:10.1128/aem.01107-08
Reinhart KO, Callaway RM (2006) Soil biota and invasive plants. New Phytol 170:445–457
Remaley T (1998) Plant Conservation Alliance, Alien Plant Working Group: Chinese Lespedeza
Richardson DM, Allsopp N, D’ANTONIO CM, Milton SJ, Rejmanek M (2000) Plant invasions: the role of mutualisms. Biol Rev 75:65–93
Rincon-Rosales R, Lloret L, Ponce E, Martinez-Romero E (2009) Rhizobia with different symbiotic efficiencies nodulate Acaciella angustissima in Mexico, including Sinorhizobium chiapanecum sp nov which has common symbiotic genes with Sinorhizobium mexicanum. FEMS Microbiol Ecol 67:103–117. doi:10.1111/j.1574-6941.2008.00590.x
Rodriguez-Echeverria S (2010) Rhizobial hitchhikers from Down Under: invasional meltdown in a plant-bacteria mutualism? J Biogeogr 37:1611–1622. doi:10.1111/j.1365-2699.2010.02284.x
Rodríguez-Echeverría S, Crisóstomo JA, Freitas H (2007) Genetic diversity of rhizobia associated with Acacia longifolia in two stages of invasion of coastal sand dunes. Appl Environ Microbiol 73:5066–5070
Rodríguez-Echeverría S, Crisóstomo JA, Nabais C, Hí F (2009) Belowground mutualists and the invasive ability of Acacia longifolia in coastal dunes of Portugal. Biol Invasions 11:651–661
Rodriguez-Echeverria S, Fajardo S, Ruiz-Diez B, Fernandez-Pascual M (2012) Differential effectiveness of novel and old legume-rhizobia mutualisms: implications for invasion by exotic legumes. Oecologia 170:253–261. doi:10.1007/s00442-012-2299-7
Sanger F, Coulson AR (1975) A rapid method for determining sequences in DNA by primed synthesis with DNA polymerase. J Mol Biol 94:441–448
Scheiner SM, Gurevitch J (eds) (1993) Design and Analysis of Ecological Experiments. Chapman & Hall, New York
Schutzenhofer MR, Knight TM (2007) Population-level effects of augmented herbivory on Lespedeza cuneata: implications for biological control. Ecol Appl 17:965–971
Smith MD, Knapp AK (2001) Physiological and morphological traits of exotic, invasive exotic, and native plant species in tallgrass prairie. Int J Plant Sci 162:785–792
Thrall PH, Slattery JF, Broadhurst LM, Bickford S (2007) Geographic patterns of symbiont abundance and adaptation in native Australian Acacia-rhizobia interactions. J Ecol 95:1110–1122. doi:10.1111/j.1365-2745.2007.01278.x
van der Heijden MGA, Klironomos JN, Ursic M, Moutoglis P, Streitwolf-Engel R, Boller T, Wiemken A, Sanders IR (1998) Mycorrhizal fungal diversity determines plant biodiversity, ecosystem variability and productivity. Nature 396:69–72
van der Putten WH (2010) Impacts of soil microbial communities on exotic plant invasions. Trends Ecol Evol 25:512–519
van der Putten WH, Klironomos JN, Wardle DA (2007) Microbial ecology of biological invasions. ISME J 1:28–37
Weir BS, Turner SJ, Silvester WB, Park DC, Young JM (2004) Unexpectedly diverse Mesorhizobium strains and Rhizobium leguminosarum nodulate native legume genera of New Zealand, while introduced legume weeds are nodulated by Bradyrhizobium species. Appl Environ Microbiol 70:5980–5987
Wilson DO (1988) Differential plant response to Inoculation with two VA mycorrhizal fungi isolated from a low-pH Soil. Plant Soil 110:69–75. doi:10.1007/Bf02143541
Wolfe BE, Klironomos JN (2005) Breaking new ground: soil communities and exotic plant invasion. Bioscience 55:477–487
Woods TM, Hartnett DC, Ferguson CJ (2009) High propagule production and reproductive fitness homeostasis contribute to the invasiveness of Lespedeza cuneata (Fabaceae). Biol Invasions 11:1913–1927
Wu TH, Hao WY, Lin XG, Shi YQ (2002) Screening of arbuscular mycorrhizal fungi for the revegetation of eroded red soils in subtropical China. Plant Soil 239:225–235. doi:10.1023/A:1015078207757
Yanagi M, Yamasato K (1993) Phylogenetic analysis of the family Rhizobiaceae and related bacteria by sequencing of 16S rRNA gene using PCR and DNA sequencer. FEMS Microbiol Lett 107:115–120
Yannarell AC, Busby RR, Denight ML, Gebhart DL, Taylor SJ (2011) Soil bacteria and fungi respond on different spatial scales to invasion by the legume Lespedeza cuneata. Front Microbiol 2:1–12. doi:10.3389/fmicb.2011.00127
Acknowledgments
The authors acknowledge A. Leonard and J. Proffitt for assistance with field work. S. Taylor, J. Dawson, K. Heath, and A. Kent provided valuable assistance during design of this research and the preparation of this manuscript. This work was supported by grant Army W9132T-09-2-0011 from the U.S. Army Engineer Research and Development Center and also by the Cooperative State Research, Education and Extension Service, U.S. Department of Agriculture, under project number ILLU-875-317.
Author information
Authors and Affiliations
Corresponding author
Additional information
Responsible Editor: Hans Lambers.
Electronic supplementary material
Below is the link to the electronic supplementary material.
ESM 1
(DOCX 91 kb)
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution License which permits any use, distribution, and reproduction in any medium, provided the original author(s) and the source are credited.
About this article
Cite this article
Hu, L., Busby, R.R., Gebhart, D.L. et al. Invasive Lespedeza cuneata and native Lespedeza virginica experience asymmetrical benefits from rhizobial symbionts. Plant Soil 384, 315–325 (2014). https://doi.org/10.1007/s11104-014-2213-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11104-014-2213-7