Abstract
The heavy metal hyperaccumulator Thlaspi caerulescens expresses several ZIP-like genes at higher levels than their orthologues in non-hyperaccumulator species, but it is not clear why. To elucidate the function of the T. caerulescens orthologues of the Arabidopsis thaliana ZIP5 and ZIP6 genes, full-length cDNAs of TcZNT5-LC and TcZNT6-LC were cloned, their expression was examined and genes were expressed in A. thaliana. Transcript level analysis revealed the constitutively high expression of these two genes in T. caerulescens compared to AtZIP5 and AtZIP6 genes and differential expression of both genes when comparing two accessions of T. caerulescens with different metal accumulation properties. Expression of TcZNT5-LC in A. thaliana did not modify Cd or Zn tolerance, but mildly affected the root and shoot Zn concentrations towards a hyperaccumulator shoot to root concentration ratio. A. thaliana zip5 knock-out mutants showed increased tolerance to Cd and decreased seed mineral concentrations. Expression of TcZNT6-LC enhanced the Cd sensitivity of A. thaliana, but no phenotype was observed for the zip6 mutant. In conclusion, the changes in expression of TcZNT5-LC and TcZNT6-LC upon changes in Zn or Cd exposure indicate both genes act in metal homeostasis, but their CaMV 35S-mediated expression in A. thaliana does not create T. caerulescens hyperaccumulator phenotypes.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Thlaspi caerulescens, a Zn/Cd/Ni hyperaccumulator, has been used as a model species for dissection of the genetic and molecular mechanism of metal accumulation and homeostasis (Assunção et al. 2003a). Hyperaccumulation of metals requires an adapted metal homeostasis allowing enhanced tolerance to metals and their accumulation. Therefore, functional and/or transcriptional modification of metal transporters, metal chelators and other proteins are needed to maintain a balance between plant metal uptake, distribution and storage (reviewed by Clemens 2001; Mäser et al. 2001; Cobbett and Goldsbrough 2002).
Members of the ZRT-IRT-like Protein (ZIP) family were the first metal transporters to be identified in plants (Eide et al. 1996). Fifteen ZIP genes have been identified in A. thaliana so far, based on whole genome sequencing. In general the ZIP transporter proteins have the capacity to transport a variety of divalent cations including Zn2+, Fe2+, Mn2+ and Cd2+ (Guerinot 2000) and the expression of ZIP genes is regulated by plant metal status due to environmental metal levels (Grotz et al. 1998). AtIRT1 and AtIRT2 have been described to have an essential role in maintenance of iron homeostasis (Connolly et al. 2002; Henriques et al. 2002; Varotto et al. 2002; Vert et al. 2001, 2002). AtZIP1, AtZIP2 and AtZIP3 confer Zn uptake activity when expressed in yeast (Grotz, et al. 1998). Genes encoding ZIPs have also been identified and characterized from other plant species, including soybean, Medicago truncatula, Thlaspi caerulescens and T. japonica (Assunção et al. 2001; Burleigh et al. 2003; Mizuno et al. 2005; Moreau et al. 2002; Pence et al. 2000; Plaza et al. 2007). TcZNT1 and TcZNT2, the proposed T. caerulescens orthologues of the A. thaliana AtZIP4 and AtIRT3 genes respectively, are constitutively over-expressed in roots (Assunção et al. 2001; Pence et al. 2000). TcZNT1 was shown to mediate high-affinity Zn uptake and low-affinity Cd uptake when expressed in yeast (Pence et al. 2000). Homologous genes TjZNT1 and TjZNT2 isolated from the Ni hyperaccumulator T. japonica were shown to be able to transport Zn, Cd and Mn (Mizuno et al. 2005).
Global transcriptomic studies revealed similar expression levels of ZIP5 and ZIP6 in roots of the Zn/Cd hyperaccumulator A. halleri and the non-hyperaccumulator A. thaliana (Weber et al. 2004). However, ZIP6 in shoots was substantially higher expressed in A. halleri than in A. thaliana (Becher et al. 2004). Expression of ZIP5 was up regulated in roots of A. thaliana upon Zn deficiency. Expression of the T. caerulescens ZIP5 orthologue (TcZNT5) was higher compared with A. thaliana (van de Mortel et al. 2006). TcZNT5 was previously cloned from T. caerulescens accession Prayon (Plaza et al. 2007). This gene was mainly expressed in roots with a much higher expression in Prayon than in the Ganges accession, which accumulates more Cd than the former (Plaza et al. 2007). These studies indicate that ZIP5 and ZIP6 and their orthologues are likely to have a role as possible Zn/Cd transporters in Zn homeostasis and Zn/Cd hyperaccumulation. Here we report the cloning of two new ZIP genes from T. caerulescens accession La Calamine (LC), which were named TcZNT5-LC and TcZNT6-LC based on their high similarity to the A. thaliana genes AtZIP5 and AtZIP6 respectively. Transcription levels of the two T. caerulescens genes were compared with their presumed A. thaliana orthologues and by comparing the T. caerulescens accessions LC and GA, which differ in Cd hyperaccumulation potential. Additional evidence for a functional role in Zn homeostasis was sought in studying T-DNA insertion knock-out mutants of AtZIP5 and AtZIP6 and in A. thaliana plants overexpressing TcZNT5-LC or TcZNT6-LC for a phenotypic change in their metal accumulation and their response to different Zn and/or Cd supply.
Materials and methods
Library screening
A cDNA library made from roots of Thlaspi caerulescens J. & C. Presl acc. La Calamine, as described by Assunção et al. (2001) was used for full-length cDNA cloning. Partial cDNA clones RR9nr066 and RR8nr089 in a pAD-GAL4-2.1 vector had been identified as putative orthologues of AtZIP5 and AtZIP6 (Rigola et al. 2006). cDNA fragments (approximately 500 bp for the ZIP5 homologue and 650 bp for ZIP6 homologue) cut from the pAD-GAL4-2.1 plasmid by EcoRI and XhoI were used as probes for cDNA library screening. The probes were labelled with [α-32P]dATP using the Hexalabel™ DNA labelling kit (Fermentas, http://www.fermentas.com/). The isolated cDNA clones were sequenced by ABI PRISM BigDye terminator cycle sequencing technology v2.0, according to the manufacturer’s instruction (Applied Biosystems; http://www.appliedbiosystems.com.), using an ABI3700 DNA analyzer. Sequence analysis was performed using the standard BLAST method (http://www.ncbi.nlm.nih.gov/BLAST/). Transmembrane domains were defined according to TMHMM (http://www.cbs.dtu.dk/services/TMHMM/). Intracellular localization was predicted using PSORT (www.psort.nibb.ac.jp). Sequence alignment was conducted by MegAlign (DNAstar, Madison WI). Phylogenetic analysis was conducted using MEGA version 3.1 (Kumar et al. 2004)
DNA blot analysis
Genomic DNA was extracted from four T. caerulescens accessions: La Calamine (LC), Monte Prinzera (MP), Ganges (GA) and an accession from Hochobir, Austria, and four Thlaspi species: T. japonicum, T. praecox, T. minimum (three hyperaccumulators) and T. perfoliatum (a non-hyperaccumulator), using a modified CTAB method (Fulton et al. 1995). Digestion of genomic DNA, TAE gel electrophoresis, DNA blotting, prehybridization, hybridization and washing were all performed as described by Assunçaõ et al. (2001). Partial cDNA fragments digested from the pAD-GAL4-2.1 vector plasmids containing full-length cDNAs using EcoRI and XhoI for TcZNT5-LC (1,032 bp) and TcZNT6-LC (1,310 bp) were used as probes and labelled as described above.
Plant materials
Seeds of A. thaliana accession Columbia-0 (Col) and T. caerulescens accession LC were germinated on garden peat soil (Jongkind BV, The Netherlands). Three-week-old seedlings were transferred to hydroponics, three plants per pot filled with 1 L modified half-strength Hoagland nutrient solution (van de Mortel et al. 2006). After growth for 3 week on this solution, the T. caerulescens plants were transferred to the same nutrient solution with a deficient (0 μM), sufficient (100 μM), or high (1,000 μM) ZnSO4 concentration on which they were grown for 7 additional days. The A. thaliana plants were transferred to the same nutrient solution with deficient (0 μM), sufficient (2 μM) or high (25 μM) ZnSO4. The nutrient solution was renewed once a week during the first 3 week and thereafter twice a week. Germination and culture were performed in a climate chamber (20/15°C day/night temperature, 12 h (A. thaliana) or 14 h days (T. caerulescens).
For comparison of transcription levels of TcZNT5 and TcZNT6 between T. caerulescens accessions LC and GA, plants were grown as described above including two additional cadmium treatments (Zn 100 μM + Cd 1 μM and Zn 100 μM + Cd 10 μM).
Semi-quantitative RT-PCR
Shoots or roots of one pot containing three A. thaliana or three T. caerulescens plants per treatment were pooled and homogenized in liquid nitrogen. Total RNA of shoots or roots was extracted with Trizol (Invitrogen) following the manufacturer’s instructions. Five micrograms of total RNA was used to synthesize cDNA with MLV reverse transcriptase (Invitrogen) and oligo (dT) as a primer (Invitrogen). The PCR amplification was performed with a 2-µl cDNA aliquot. The AtZIP5 forward primer was 5′-ATGAGAATCACACAAAACGTCAAGC-3′ and the reverse primer was 5′-TGGGATTCACCAGATTCCAC-3′; the TcZNT5 forward primer was 5′-ACCGGAGCCGAGTTGTG-3′ and the reverse primer was 5′-TGGGCCATGATTTGAAGC-3′; the AtZIP6 forward primer was 5′-GTCACCGGAACAGAGGCAGCAA-3 and the reverse primer was 5′-TTCACCGCAAGTCGTCAGCATCTT-3′; the TcZNT6 forward primer was 5′-AGAGACGGAGACGCGGCGG-3′ and the reverse primer was 5′-CTGATGAAACGAAAGAGTAGCG-3′. Primer pairs for Tubulin were used as a control for using similar cDNA quantities for each sample. For A. thaliana the forward Tubulin primer was 5′-AAGCTTGCTGATAACTGTACTGGT-3′ and the reverse primer was 5′GGTTTGGAACTCAGTGACATCA-3′; for T. caerulescens the forward primer was 5′-CTACGCACCAGTCATCTCT-3′ and the reverse primer was 5′-CGAGATCACCTCCTGGAACA-3′. For each comparative analysis several PCR cycles were run and the number of cycles which showed the best contrast between species, accessions or metal exposures was shown. For all for Tubulin amplifications 25 PCR cycles were performed for both A. thaliana and T. caerulescens samples. PCR fragments were separated on an ethidium bromide stained 1 % agarose TAE gel.
Construction of expression vectors
The full-length cDNAs of TcZNT5-LC and TcZNT6-LC were amplified from the pAD-GAL4-2.1 vector plasmids by PCR. Gateway primers were used for PCR, the forward primer was 5′-GGGGACAAGTTTGTACAAAAAAGCAGGCTGATCGAATTAGGATCCTCTGC-3′, containing the attB1 sequence (underlined) and a BamHI site (italic) in pAD-GAL4-2.1, and the reverse primer was 5′- GGGGACCACTTTGTACAAGAAAGCTGGGTCTAATGGGCTCGAGAGTCGAC-3′, containing the attB2 sequence (underlined) and an XhoI site (italic) in pAD-GAL4-2.1. The PCRs were performed with the proofreading Pfu polymerase (Fermentas, http://www.fermentas.com) at 93°C for 5 min, followed by 30 cycles of 93°C for 1 min, 55°C for 1 min and 68°C for 3 min, and finished by an extension at 68°C for 10 min. PCR products were recombined into pDONR207 (Invitrogen, http://www.invitrogen.com) in a 10-μl BP Clonase (Invitrogen) reaction following the manufacturer’s instruction. The fragments were transferred from their donor constructs to the binary over-expression vector pGD625 (de Folter et al. 2006), under the control of the double 35S CaMV enhancer, in a 10-μl LR Clonase (Invitrogen) reaction following the manufacturer’s instruction. The binary constructs were introduced into Agrobacterium tumefaciens strain AGL0 by electroporation.
Plant transformation and metal tolerance/sensitivity growth assays
The binary constructs with full-length cDNAs of TcZNT5-LC and TcZNT6-LC were used to transform A. thaliana Col by the standard flower dip method (Clough and Bent 1998). The primary T1 transformants were selected on half-strength MS medium containing kanamycin (50 mg/ml). Kanamycin-resistant plants were transferred to soil, and the T2 seeds resulting from self-fertilization were collected. The T2 seeds were plated on the same selection medium and scored for kanamycin resistance. Transgenic lines that displayed a 3:1 segregation ratio for kanamycin resistance to sensitivity in the T2 generation indicating a single T-DNA insertion locus were selected for further analysis. In the T3 generation, we tested 12 T3 progeny plants from ten T2 plants each by PCR using the gene specific primers for TcZNT5 and TcZNT6 designed for the semi-quantitative RT-PCR. Genomic DNA was isolated using a modified CTAB method (Fulton et al. 1995). The lines for which all the 12 T3 progenies contained a T-DNA were deemed to be homozygous transgenics. T4 seeds from these plants were used for further experiments. The expression levels of TcZNT5-LC and TcZNT6-LC in the transgenic lines were determined by RT-PCR using total RNA extracted from T4 plants grown in soil for 30 days.
Selection of homozygous T-DNA insertion plants
SALK_009007 and SALK_116013 A. thaliana T-DNA insertion lines were obtained from the Nottingham Arabidopsis Stock Centre (NASC). SALK_009007 carries a T-DNA insert approximately 357 bp downstream of the start of the first exon of AtZIP5 (At1g05300), while SALK_116013 carries a T-DNA insert approximately 363 bp downstream of the start of the first exon of AtZIP6 (At2g30080) (Alonso et al. 2003). Genomic DNA was isolated from eight plants of SALK_009007 and 16 plants of SALK_116013. Homozygous mutants were identified by PCR using genomic DNA as template and separated combinations of primer LBb1, designed to fit the left border of the T-DNA insert (5′- GCGTGGACCGCTTGCTGCAACT-3′) (http://signal.salk.edu), and AtZIP5 or AtZIP6 specific primer pairs. The AtZIP5 gene specific primers and the AtZIP6 forward primer were the same as used in the semi-quantitative RT-PCR. The reverse primer for AtZIP6 was 5′- TGCAACCACCAAGACCCAAA-3′. Seeds harvested from the identified homozygous T-DNA insertion plants and homozygous wild-type plants segregating in the respective SALK lines, were used for further phenotype screening.
Metal tolerance screening
Seeds were sterilized and sown on half-strength MS medium containing deficient or high Zn (0 μM or 200 μM), deficient Fe (0.5 μM), Cd (50 μM) and Fe deficient plus Cd (0.5 μM Fe + 50 μM Cd). Half-strength MS medium was used as control. For each treatment, five replicates were used for each line with 20 seeds sown in each replicate. After being sown on media, seeds were kept at 4°C for 4 days in the dark to synchronize germination. Seedlings were grown in a 14 h light, 10 h dark cycle at 20°C day and 15°C night temperatures. The phenotypes were studied on 15-day-old plants by measuring root length.
Metal content measurement
Shoot metal concentrations were determined for wild-type A. thaliana, the overexpressing lines and T-DNA insertion mutant lines, with five replicates for each line. Plants were grown for 3 weeks in hydroponic solution (1/10 Hoagland’s) containing 1 μM Zn, Cd and Ni. The solution was pH-buffered at 5.6 with 2 mM MES (2-morpholino-ethanesulphonic acid). Plants were rinsed with deionised water, separated into roots and shoots. Roots were then placed in an ice-cold solution of 1 mM LaCl3 and 5 mM CaCl2 for 20 min to desorb apoplastic metals. Roots and shoots were blotted dry with tissue paper and weighed. Roots and shoots were digested with 5 ml high purity HNO3/HClO4 (85:15, v:v). Metal concentrations were determined by inductively coupled plasma mass spectrometry (ICP-MS; Agilent ICP-MS 7500ce, Agilent Technologies, Santa Clara, CA, US).
The seed mineral concentrations were measured for wild type A. thaliana Col and zip5 mutant plants. Seedlings were grown on a standard hydroponics solution suggested for A. thaliana (Tocquin et al. 2003), in a phytotron at 20°C with a relative humidity of 70% and a light intensity of 40 W/m2 for 12 h/day. Seeds were harvested when completely mature and dry. Seed mineral concentrations were determined by Atomic Absorption Spectrometry (Perkin Elmer AAS 1100; Perkin Elmer, Rodgau-Judesheim, Germany) after acid digestion.
Statistical analysis
The phenotype data were analyzed by analysis of variance. The significance of the difference between mutant or over-expression lines with wild-type Col was determined by the use of least significant difference (LSD) at the 5% probability level.
Results
Cloning of full-length cDNA clones for TcZNT5-LC and TcZNT6-LC
Two partial T. caerulescens cDNA clones (RR9nr066 and RR8nr089), previously identified to have high DNA identity respectively to AtZIP5 (At1g05300) and AtZIP6 (At2g30080) (Rigola et al. 2006), were used as probes to screen a cDNA library prepared from roots of T. caerulescens accession La Calamine (LC) (Assunção et al. 2001). Three positive clones were obtained for each probe. One of the three positive clones detected with the RR9nr066 probe contained an open reading frame (ORF) of 1,068 bp covering the full predicted AtZIP5 ORF and encoding a predicted protein of 355 amino acids and 37 kDa. It contains eight transmembrane domains (TMs) (Supplemental Fig. 1). The sequence has 86% DNA identity and 90% amino acid identity with AtZIP5 and was named TcZNT5-LC. There is a putative metal binding domain containing a series of repeated histidine residues (HVHAHGHAHG) between TM4 and TM5, and it also has the conserved histidine residue in TM5 found in many members of the ZIP family. According to TAIR (www.arabidopsis.org), both AtZIP5 and the related protein AtZIP3 are targeted to the plasma membrane. Analysis by PSORT also predicted the TcZNT5-LC protein to be targeted to the plasma membrane. There are three other full-length cDNA sequences of TcZNT5 from other accessions of T. caerulescens, one for GA and two for Prayon (PR), deposited at NCBI GenBank (AJ937738 (GA) and AJ937739 and AF292029 (PR)). The TcZNT5-LC and TcZNT5-PR amino acid sequences are identical, while the predicted protein encoded by TcZNT5-GA lacks three amino acids close to the N-terminus, compared to TcZNT5-LC and TcZNT5-PR.
The sequence of the longest of three positive clones identified by the RR8nr089 probe contains an ORF of 1,023 bp encoding a predicted protein of 340 amino acids, with a molecular weight of 35 kDa. The protein contains eight potential TMs (Supplemental Fig. 2) and is also predicted to be targeted to the plasma membrane by PSORT. The cDNA sequence has 86% DNA identity and 92% amino acid identity with AtZIP6 and therefore the sequence was named TcZNT6-LC. The predicted TcZNT6-LC amino acid sequence contains the conserved histidine residue in TM4, but in contrast to most members of the ZIP family, TcZNT6-LC contains only two histidine residues as potential metal binding motifs in the variable region between TM3 and TM4.
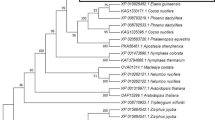
The predicted amino acid sequences of TcZNT5-LC and TcZNT6-LC were aligned with other members of the ZIP gene family. Phylogenetic analysis (Fig. 1) revealed that TcZNT5-LC is closest to AtZIP5 as expected based on BLAST analysis, and both are closely related to AtZIP3. TcZNT6-LC is closest to AtZIP6 and both are more distantly related to the other ZIP family members. According to the previous studies on ZIP family members in T. caerulescens and the nomenclature used before (Pence et al. 2000; Assunção et al. 2001), we propose to maintain the provisional names for these two genes as TcZNT5 and TcZNT6.
Phylogenetic comparison of predicted protein sequences of TcZNT5 and TcZNT6 with those of 22 ZIP family members from Arabidopsis thaliana, A. halleri, Thlaspi caerulescens and T. japonicum. The phylogenetic analysis was conducted using MEGA version 3.1 (Kumar et al. 2004). GenBank accession numbers are: AtIRT1 (NM_118089), AtIRT2 (NM_001036593), AtIRT3 (NM_104776), AtZIP1 (NM_112111), AtZIP2 (NM_125344), AtZIP3 (NM_128786), AtZIP4 (NM_100972), AtZIP5 (NM_100409) AtZIP6 (NM_128563), AtZIP7 (NM_126440), AtZIP8 (NM_148089), AtZIP9 (NM_119456), AtZIP10 (NM_102864), AtZIP11 (NM_104468), AtZIP12 (NM_125609), AhZIP6 (AJ580315), TcZNT1 (AF275751), TcZNT2 (AF275752), TcZNT5-PR (AF292029), TcZNT5-GA (AJ937739), TjZNT1 (AB206397), TjZNT2 (AB175740). The ruler indicates genetic distance as the number of substitutions per unit time
DNA blot analysis was performed to determine the copy number of the TcZNT5 and TcZNT6 genes in the T. caerulescens genome and in those of related Thlaspi species (Fig 2). We used restriction enzymes EcoRI (only for TcZNT5) and XbaI for genomic DNA digestion; EcoRI cuts once in the TcZNT5-LC cDNA sequence, XbaI does not cut in the TcZNT5-LC or TcZNT6-LC cDNA sequences. In all T. caerulescens accessions and other Thlaspi species tested, there were at least two bands for the EcoRI and XbaI digestions hybridized with the TcZNT5-LC probe (Fig. 2a), one strong band and one or two low intensity bands, suggesting additional gene copies with lower similarity to the probe next to the ZNT5 copies. As was previously found for NRAMP genes (Oomen et al. 2008), there is much stronger hybridization of the TcZNT5-LC probe to the lanes with T. japonicum genomic DNA than to other lanes. As the amount of genomic DNA loaded for T. japonicum was comparable to that of T. caerulescens (data not shown), it appears that multiplications of the TjZNT5 gene have occurred in this species.
DNA blot analysis of ZNT5 and ZNT6 in T. caerulescens accessions and Thlaspi species. Genomic DNA was digested with EcoRI or XbaI. DNA blots were hybridized with a TcZNT5 (a) or TcZNT6 (b) cDNA fragment probe. The numbers above the lanes designate different T. caerulescens accessions: La Calamine (1), Monte Prinzera (2), Ganges (3), Hochobir (7); or Thlaspi species: T. japonica (4), T. praecox (5), T. minimum (6) and T. perfoliatum (8). Size markers at shown on the left
For TcZNT6 some T. caerulescens accessions showed only one hybridizing fragment (Fig 2b), in accordance with the presence of only one copy of this gene in this species. TjZNT6 is not multiplied to the same extent as TjZNT5 in T. japonicum.
Expression of TcZNT5 and TcZNT6 in T. caerulescens
The role of TcZNT5 and TcZNT6 in metal accumulation and homeostasis in T. caerulescens (accession LC) was first evaluated by examining their expression upon exposure to various metal conditions. Transcript levels of TcZNT5 and TcZNT6 in shoots and roots were compared to those of their AtZIP5 and AtZIP6 orthologues in A. thaliana using semi-quantitative RT-PCR on 4-week-old, hydroponically grown plants. In general, expression of TcZNT5 and TcZNT6 is higher than that of AtZIP5 and AtZIP6, both in roots and shoots (Fig. 3). Expression of AtZIP5 was induced under Zn deficiency, whereas expression of AtZIP6 was induced by high Zn supply. The transcript level of TcZNT5 was higher in roots than in shoots, while TcZNT6 was expressed at a similar level in shoots and roots under the Zn levels tested. In A. thaliana, AtZIP5 is expressed at a similar level in roots and shoots, while AtZIP6 expression is higher in roots than in shoots.
Semi-quantitative RT-PCR analysis of T. caerulescens TcZNT5 and TcZNT6 and their A. thaliana orthologues AtZIP5 and AtZIP6. Shoots and roots were used from plants grown at 0, 100 and 1,000 µM Zn for T. caerulescens and 0, 2 and 25 µM Zn for A. thaliana. RT-PCR was performed using specific primer pairs for each gene. RT-PCR (30 cycles) was used to amplify a 424-bp fragment for TcZNT5, a 605-bp fragment for TcZNT6, a 593-bp fragment for AtZIP5 and a 679-bp fragment for AtZIP6. Tubulin amplification (25 cycles) was used as control for equal use of cDNA
As T. caerulescens is a Zn/Cd hyperaccumulator, we further tested the changes of transcript levels of TcZNT5 and TcZNT6 in roots and shoots upon exposure to deficient (0 μM), sufficient (10 μM and 100 μM) and high Zn (1,000 μM) conditions or with sufficient Zn (100 μM) in the presence of low Cd (1 μM) or high Cd (10 μM) concentrations in two T. caerulescens accessions, LC and GA. These two accessions are similar in Zn/Cd tolerance, but under the conditions we used, GA accumulates more Zn and Cd than LC (Assunção et al. 2003b). As shown before, TcZNT5 was predominantly expressed in roots in both accessions (Fig. 4), but with a higher expression in LC compared to GA. Although much less than what we observed for AtZIP5, expression of TcZNT5 was also slightly up-regulated in roots under Zn deficiency in both accessions. Cd treatment did not alter the expression level of TcZNT5 in LC or GA roots or leaves. There was a difference in expression of TcZNT6 between two accessions in roots. TcZNT6 was expressed at similar levels in shoots and roots in GA, irrespective of the metal supply. In LC the expression in shoots was the same, but in roots expression was lower than in GA and down-regulated in response to 10 µM Cd (+100 µM Zn).
Zn and Cd exposure effects on TcZNT5 and TcZNT6 transcript levels in T. caerulescens accessions Ganges and La Calamine. Shoots and roots were used from plants grown at 0, 100 and 1,000 µM Zn, or 100 µM Zn supplemented with 1 or 10 µM Cd. RT-PCR was performed (25 cycles) using gene specific primers to amplify a 424-bp fragment from TcZNT5 and 605-bp fragment from TcZNT6. Tubulin amplification (25 cycles) was used as a control of equal cDNA use
Phenotypic analysis of Arabidopsis zip5 T-DNA insertion mutants and TcZNT5-LC expressing plants
Investigating the function of TcZNT5 and TcZNT6 in T. caerulescens is cumbersome in the absence of a collection of T-DNA insertion mutants like in A. thaliana. Therefore we analyzed T-DNA insertion mutants of their orthologues, AtZIP5 and AtZIP6, in A. thaliana. Homozygous zip5 T-DNA insertion plants and wild-type (WT) sister plants were isolated by PCR screening (data not shown). RT-PCR confirmed the expression of ZIP5 in the WT plants. No expression was detected in the mutant (data not shown) indicating that the T-DNA is not spliced from the transcript and that the insertion is likely to cause a loss-of-function null allele.
In addition the TcZNT5-LC full-length cDNA was expressed under control of the constitutive CaMV 35S promoter in A. thaliana Col. Three independent homozygous single-locus transgenic expression lines were obtained. The TcZNT5-LC transgene expression was higher in line no. 6 than in the other two lines, as found by semi-quantitative RT-PCR (Fig. 5).
Semi-quantitative RT-PCR analysis of TcZNT5-LC or TcZNT6-LC expression of wild-type (WT) and homozygous transgenic A. thaliana lines transformed with a CaMV 35S::TcZNT5-LC (ZNT5) or CaMV 35S::TcZNT6-LC (ZNT6) construct. Tubulin amplification was used as a control of equal cDNA use. The number of PCR cycles is indicated
When grown in soil, both zip5 mutants and transgenic plants expressing TcZNT5-LC showed no visible morphological differences compared to WT plants. When screened for root growth on vertical plates, zip5 seedlings had 19.3% longer roots than WT seedlings when grown on medium containing 50 μM Cd (P < 0.05), suggesting increased Cd tolerance. However this difference disappeared when plants were grown on medium with low Fe supply (0.5 μM) in the presence of Cd (Fig. 6a). In contrast, no significant differences in root lengths were detected for TcZNT5-LC expressing lines compared with WT at low Zn, low Fe and high Cd supply (Fig. 6b).
A. thaliana zip5 T-DNA insertion loss-of-function mutants and transgenic lines expressing TcZNT5-LC, showed phenotypic changes in tolerance and mineral accumulation. Average root length (± SE) of 15-day-old A. thaliana wild-type (WT) and zip5 seedlings (a) or 35S::TcZNT5-LC expression lines nos. 2, 4 and 6 (b), germinated and grown on half-strength MS medium containing different concentrations of Zn, Fe and Cd. Five replicates with each 12 to 20 seedlings were measured for each treatment. c Mineral concentrations (± SE) in zip5 mutant roots (zip5R) and shoots (zip5S) or in wild-type roots (WTR) and shoots (WTR) of hydroponically grown plants (1/10 Hoagland’s solution) supplied with 100 μM Fe-EDTA and 1 μM Zn, Cd and Ni. d Seed Zn, Mn, Fe, K and Ca concentrations (± SE) of wild-type (WT) and zip5 mutant plants. e Average number of seeds per fruit (SNF), fruit length (FL, in cm) and seed weight (SW, in mg per 100 seeds) of WT and zip5 mutant plants. (f) and (g) Mineral concentrations (± SE) in roots (f) and shoots (g) of hydroponically grown (1/10 Hoagland’s solution containing 100 μM Fe-EDTA and 1 μM Zn, Cd and Ni) 14-day-old seedlings of 35S::TcZNT5-LC expression lines nos. 2, 4 and 6. For (c), (f) and (g) we divided the actual root Fe concentrations by 100 and the actual root Zn and root and shoot Cd concentrations by 10 in order to show all mineral concentrations relative to each other in the same graphs. * Significantly different at P < 0.05. Significance is determined by one-way ANOVA. Five replicates were measured for each treatment in (c), (f) and (g)
Mineral concentrations were determined in roots, shoots and seeds of the zip5 mutant plants and roots and shoots of A. thaliana lines expressing TcZNT5-LC grown in hydroponics. Shoot Fe and Zn concentrations were significantly higher in zip5 (27 % and 18 %, respectively), while root Cu and Cd concentrations were significantly lower (55% and 26%, respectively) compared with WT (P < 0.05) (Fig. 6c). Seed Fe, K, Mg and P concentrations were significantly decreased in zip5 (Fig. 6d). An evaluation of growth related traits also showed that fruit length and seed weight were a little, but significantly, decreased in zip5 (Fig. 6e), indicating that inactivation of ZIP5 disturbs the normal seed setting of A. thaliana plants. Plants expressing TcZNT5-LC did not show a univocally strong change in mineral accumulation. However, two of the three transgenic lines (no. 4 and no. 6) showed a significant decrease in root Zn concentrations, 40% and 78% of WT (Fig. 6f), and the line with higher expression of TcZNT5-LC (no. 6) also showed a significantly enhanced shoot Zn concentration (50% higher) when compared to wild type (P < 0.05) (Fig. 6g). Consequently their shoot to root Zn concentration ratios increased, more resembling T. caerulescens ratios. The other transgenic line (no. 2) showed a significant increase in the root Cd concentration (P < 0.05) (Fig. 6f).
Phenotypic analysis of Arabidopsis zip6 T-DNA insertion mutants and TcZNT6-LC expressing plants
A. thaliana T-DNA insertion mutants of AtZIP6 and transgenic lines expressing TcZNT6-LC were analyzed for their phenotype differences compared with WT. Three independent homozygous single-locus transgenic expression lines were studied. One of the three lines (no. 14) showed a high expression level of the transgene, the other two lines showed lower expression of the transgene, but still higher than expected for the AtZIP6 gene (Fig. 5).
Like zip5 mutants and TcZNT5-LC expressors, zip6 mutants and plants expressing TcZNT6-LC did not show obvious morphological alterations compared to WT when they were grown in soil. When screened for root growth on vertical plates, no significant differences in root length were detected when comparing zip6 mutants with WT plants, irrespective of the metal exposures (Fig. 7a). However, for transgenic lines, high expression of TcZNT6-LC (no. 14) resulted in significantly reduced root length compared to the WT plants and plants with modest TcZNT6-LC expression (no. 11 and no. 18) on medium supplemented with Cd (50 μM) (P < 0.001), both under sufficient and deficient Fe supply, thus enhancing the Cd sensitivity (Fig. 7b). The exposures to Zn deficiency or excess Zn showed no significant differences in root length.
zip6 T-DNA insertion loss-of-function mutants and transgenic A. thaliana plants expressing TcZNT6-LC showed phenotypic changes in tolerance and mineral accumulation. Average root length (± SE) of 15-day-old A. thaliana wild-type (WT) and zip6 seedlings (a) or 35S::TcZNT6-LC expression lines nos. 11, 14 and 18 (b), germinated and grown on half-strength MS medium containing different concentrations of Zn, Fe and Cd. Five replicates with each 12 to 20 seedlings were measured for each treatment. * indicates significantly different at P < 0.001 as determined by one-way ANOVA. Five replicates, each with 12 to 20 seedlings, were measured for each treatment. c Mineral concentrations (± SE) in zip6 mutant roots (zip6R) and shoots (zip6S) or in wild-type roots (WTR) and shoots (WTR) of hydroponically grown plants (1/10 Hoagland’s solution) supplied with 100 μM Fe-EDTA and 1 μM Zn, Cd and Ni. d and e Mineral concentrations (± SE) in roots (d) and shoots (e) of hydroponically grown (1/10 Hoagland’s solution containing 100 μM Fe-EDTA and 1 μM Zn, Cd and Ni) 14-day-old seedlings of 35S::TcZNT6-LC expression lines nos. 11, 14 and 18. For c, d and e we divided the actual root Fe concentrations by 100 and the actual root Zn and root and shoot Cd concentrations by 10 in order to show all mineral concentrations relative to each other in the same graphs. * Significantly different at P < 0.05. Significance is determined by one-way ANOVA. Five replicates were measured for each treatment in (c), (d) and (e)
Mineral concentrations were determined in roots and shoots of zip6 mutant plants and TcZNT6-LC expressing plants grown in hydroponics. The root Fe concentration was significantly lower (17%) in zip6 plants than in WT plants, but the shoot Mn concentration was higher (25%) in zip6 than in WT (Fig. 7c) (P < 0.05). The three independent TcZNT6-LC expressing lines all showed significantly higher shoot Mn concentrations compared with WT (P < 0.05) (Fig. 7d). Only line 11 showed a significantly higher Fe concentration in roots (P < 0.05) (Fig. 7e).
Discussion
Screening of a T. caerulescens root cDNA library identified full-length cDNA clones of two ZIP family members, TcZNT5-LC and TcZNT6-LC, the latter of which had not been identified in T. caerulescens before. The predicted proteins encoded by the two genes share all common features found in ZIP proteins: they have eight putative transmembrane domains (TM); they contain a histidine repeat in the variable region, which has been proposed as the metal binding and/or sensing site (Grossoehme et al. 2006); and they have a conserved histidine residue in TM4 or TM5, which is predicted to occupy the polar face of the amphipathic helix and has a role in substrate transport through the membrane (Eng et al. 1998; Guerinot 2000).
The N-termini of the predicted protein sequences of the four identified TcZNT5 alleles from accessions LC, GA and PR (two alleles) are different from the predicted protein sequence of the A. thaliana AtZIP5 gene. All T. caerulescens alleles appear to lack one of the codons encoding a phenylalanine residue. However, when comparing the AtZIP5 predicted protein sequence to that of the related gene AtZIP3, it shows that AtZIP3 is also lacking the phenylalanine residue and an adjacent leucine residue, suggesting that the phenylalanine residue may not be critical in terms of providing proper protein function. Also among the four T. caerulescens alleles there are some differences. Most striking is the absence of three codons encoding Lys8-Leu9-Leu10 in the predicted TcZNT5-GA protein sequence. It will be interesting to determine if this difference affects the subcellular localization of the TcZNT5 protein.
Both TcZNT5 and TcZNT6 are more or less constitutively expressed in T. caerulescens La Calamine, with both root and shoot expression staying fairly constant over a wide range of Zn supplies (0–1,000 μM). This is different from the Zn-status-dependent regulation of the expression of their orthologues AtZIP5 and AtZIP6 in A. thaliana (Fig 3). Both TcZNT5 and TcZNT6 are also expressed at a higher level in T. caerulescens than their orthologues in A. thaliana. The RT-PCR results presented here confirm the root micro-array expression data for TcZNT5 and AtZIP5 as determined by van de Mortel et al. (2006). They did not report a higher root expression of TcZNT6 compared to AtZIP6 as the difference did not exceed the 5-fold threshold level used as a cut-off in that analysis. A similarly high and constitutive expression was previously reported for the TcZNT1 and TcZNT2 genes in T. caerulescens and the TgMTP1 and AhMTP1 genes in the metal hyperaccumulator species T. goesingense and A. halleri (Assunção et al. 2001; Dräger et al. 2004; Pence et al. 2000; Persans et al. 2001). As Zn hyperaccumulation in T. caerulescens is a constitutive trait involving strongly enhanced metal uptake and root to shoot translocation (Assunção et al. 2003a), higher cation transport capacity will be needed for taking up metals from soil and for intracellular storage and allocation (Lasat et al. 1996). T. caerulescens seems to have dealt with the demand for higher transport capacity by increasing the expression of the available zinc transporter genes.
The similarities in protein sequence between TcZNT5 in T. caerulescens and AtZIP5 in A. thaliana and the predominant expression of TcZNT5 in roots provide evidence for a presumed function of TcZNT5 in cellular Zn uptake mainly in roots of T. caerulescens. Since AtZIP5 is expressed more or less equally in roots and shoots, it is likely to have such a function in both roots and shoots. Nevertheless, the A. thaliana knock-out zip5 mutant did not show a strong phenotype when exposed to different metal supplies, which suggests that the AtZIP5 gene function is largely redundant. This is not unexpected in view of the presence of the highly similar AtZIP3 gene. Double mutant zip3zip5 plants need to be studied to examine this further.
When comparing the two accessions of T. caerulescens, expression of TcZNT5 in roots is slightly higher in LC than in GA with or without Cd exposure. Plaza et al. (2007) also reported a higher expression of TcZNT5 in GA than in PR, an accession very similar to LC both in origin and metal accumulation characteristics (Lombi et al. 2001) and in TcZNT5 allele coding sequence. As GA is a much better Cd hyperaccumulator than LC under the investigated conditions (Assunção et al. 2003b), TcZNT5 is unlikely to play a dominant role in Cd uptake in T. caerulescens roots. Since studying the gene functions in T. caerulescens by transgenics is cumbersome, we aimed to mimic the higher expression in GA by expression of TcZNT5-LC under the control of the CaMV 35S promoter in A. thaliana. The high expression of TcZNT5-LC in A. thaliana also promoted a shift in accumulation of Zn towards leaves, although clearly not at the level of a Zn hyperaccumulator and only in two of the three expressors. Obviously only altering expression of one gene, TcZNT5, is not enough to fully mimic a hyperaccumulation phenotype and it may well be that tissue-specific expression is needed rather than the general, high ectopic expression created by the CaMV 35S promoter.
Comparison of the TcZNT6 expression in GA and LC showed a lower expression in LC roots at equal Zn supply and a clear down-regulation in LC upon increased Cd supply. Knowing that GA is a slightly better Zn hyperaccumulator and a much better Cd hyperaccumulator than LC under the conditions tested (Assunção et al. 2003b), this supports a role for this protein in shoot Zn storage and suggests some ability to transport Cd. Similar to the expression pattern of TcZNT6 in T. caerulescens LC, AhZIP6 is also more highly expressed in shoots than in roots of the Zn/Cd hyperaccumulator A. halleri (Becher et al. 2004). The main difference between A. thaliana and its hyperaccumulating relative is a higher accumulation of metals in the shoots. Therefore the stronger expression of TcZNT6/AhZIP6 in shoots and the increased expression of AtZIP6 upon increasing Zn conditions further suggest these genes may play a role in storage or detoxification of Zn, or in maintaining homeostasis of other metals like Fe or Mn, which is affected by enhanced Zn or Cd exposure, as was observed by van de Mortel et al. (2006; 2008).
In conclusion, TcZNT5 and TcZNT6, two ZIP genes cloned from T. caerulescens, are constitutively highly expressed and likely to encode metal transporters with Zn, Cd, and Fe or Mn transport ability. Additional studies are needed to further elucidate their exact role in plant mineral homeostasis and metal hyperaccumulation.
References
Alonso JM, Stepanova AN, Leisse TJ, Kim CJ, Chen H, Shinn P, Stevenson DK, Zimmerman J, Barajas P, Cheuk R, Gadrinab C, Heller C, Jeske A, Koesema E, Meyers CC, Parker H, Prednis L, Ansari Y, Choy N, Deen H, Geralt M, Hazari N, Hom E, Karnes M, Mulholland C, Ndubaku R, Schmidt I, Guzman P, Aguilar-Henonin L, Schmid M, Weigel D, Carter DE, Marchand T, Risseeuw E, Brogden D, Zeko A, Crosby WL, Berry CC, Ecker JR (2003) Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301:653–657
Assunção AGL, Martins da Costa P, de Folter S, Vooijs R, Schat H, Aarts MGM (2001) Elevated expression of metal transporter genes in three accessions of the metal hyperaccumulator Thlaspi caerulescens. Plant Cell Environ 24:217–226
Assunção AGL, Schat H, Aarts MGM (2003a) Thlaspi caerulescens, an attractive model species to study heavy metal hyperaccumulation in plants. New Phytol 159:351–360
Assunção AGL, ten Bookum WM, Nelissen HJM, Vooijs R, Schat H, Ernst W (2003b) Differential metal-specific tolerance and accumulation patterns among Thlaspi caerulescens populations originating from different soil types. New Phytol 159:411–419
Becher M, Talke IN, Krall L, Krämer U (2004) Cross-species microarray transcript profiling reveals high constitutive expression of metal homeostasis genes in shoots of the zinc hyperaccumulator Arabidopsis halleri. Plant J 37:251–268
Burleigh SH, Kristensen BK, Elldgarrd Bechmann I (2003) A plasma membrane zinc transporter from Medicaga truncatula is up-regulated in roots by Zn fertilization, yet down-regulated by arbuscular mycorrhizal colonization. Plant Mol Biol 52:1077–1088
Clemens S (2001) Molecular mechanisms of plant metal tolerance and homeostasis. Planta 212:475–486
Clough SJ, Bent A (1998) Floral dip: a simplified method for Agrobacterium—mediated transformation of Arabidopsis. Plant J 16:735–743
Cobbett C, Goldsbrough P (2002) Phytochelatins and metallothioneins: roles in heavy metal detoxification and homeostasis. Annu Rev Plant Biol 53:159–182
Connolly EL, Fett JP, Guerinot ML (2002) Expression of the IRT1 metal transporter is controlled by metals at the levels of transcript and protein accumulation. Plant Cell 14:1347–1357
de Folter S, Shchennikova AV, Franken J, Busscher M, Basker R, Grossniklaus U, Angenent GC, Immink GH (2006) A B sister MADS-box gene involved in ovule and seed development in petunia and Arabidopsis. Plant J 47:934–946
Dräger DB, Desbrosses-Fonrouge AG, Krach C, Chardonnens AN, Meyer RC, Saumitou-Laprade P, Krämer U (2004) Two genes encoding Arabidopsis halleri MTP1 metal transport proteins co-segregate with Zn tolerance and account for high MTP1 transcript levels. Plant J 39:425–439
Eide D, Broderius M, Fett J, Guerinot ML (1996) A novel iron-regulated metal transporter from plants identified by functional expression in yeast. Proc Natl Acad Sci USA 93:5624–5628
Eng BH, Guerinot ML, Eide D, Saier MH (1998) Sequence analysis and phylogenetic characterization of the ZIP family of metal ion transport proteins. Membrane Bio 166:1–7
Fulton TM, Chunwongse J, Tanksley SD (1995) Microprep protocol for extraction of DNA from tomato and other herbaceous plants. Plant Mol Biol Rep 13:207–209
Grossoehme NE, Akilesh S, Guerinot ML, Wilcox DE (2006) Metal-binding thermodynamics of the histidine-rich sequence from the metal-transport protein IRT1 of Arabidopsis thaliana. Inorg Chem 45:8500–8508
Grotz N, Fox T, Connolly E, Park W, Guerinot ML, Eide D (1998) Identification of a family of zinc transporter genes from Arabidopsis that respond to zinc deficiency. Proc Natl Acad Sci USA 95:7220–7224
Guerinot ML (2000) The ZIP family of metal transporters. BBA 1465:190–198
Henriques R, Jasik J, Klein M, Martinoia E, Feller U, Schell J, Pais MS, Koncz C (2002) Knock-out of Arabidopsis metal transporter gene IRT1 results in iron deficiency accompanied by cell differentiation defects. Plant Mol Biol 50:587–597
Kumar S, Tamura K, Nei M (2004) MEGA3: integrated software for molecular evolutionary genetics analysis and sequence alignment. Brief Bioinform 5:150–163
Lasat MM, Baker AJM, Kochian LV (1996) Physiological characterization of root Zn2+ absorption and translocation to shoots in the Zn hyperaccumulator Thlaspi caerulescens. Plant Physiol 119:305–311
Lombi E, Zhao FJ, Dunham SJ, McGrath SP (2001) Cadmium accumulation in populations of Thlaspi caerulescens and Thlaspi goesingense. New Phytol 145:11–20
Mäser P, Thomine S, Schroeder JI, Ward JM, Hirschi K, Sze H, Talke IN, Amtmann A, Maathuis FJM, Sanders D, Harper JF, Tchieu J, Gribskov M, Persans MW, Salt DE, Kim SA, Guerinot ML (2001) Phylogenetic relationships within cation transporter families of Arabidopsis. Plant Physiol 126:1646–1667
Mizuno T, Usui K, Horie K, Nosaka S, Mizuno N, Obata H (2005) Cloning of three ZIP/Nramp transporter genes from a Ni hyperaccumulator plant Thlaspi japonicum and their Ni2+- transport abilities. Plant Physiol Biochem 43:793–801
Moreau S, Thomson RM, Kaiser BN, Trevaskis B, Guerinot ML, Udvardi MK, Puppo A, Day DA (2002) GmZIP1 encodes a symbiosis-specific zinc transporter in soybean. J Biol Chem 277:4738–4746
Oomen RJFJ WuJ, Lelièvre F, Blanchet S, Barbier-Brygoo H, Aarts MGM, Thomine S (2008) Functional characterization of NRAMP3 and NRAMP4 from the metal hyperaccumulator Thlaspi caerulescens. New Phytol 181:637–650
Pence NS, Larsen PB, Ebbs S, Letham DLD, Lasat MM, Garvin DF, Kochian LV (2000) The molecular physiology of heavy metal transport in the Zn/Cd hyperaccumulator Thlaspi caerulescens. Proc Natl Acad Sci USA 97:4596–4960
Persans MW, Nieman K, Salt D (2001) Functional activity and role of cation-efflux family members in Ni hyperaccumulation in Thlaspi goesingense. Proc Natl Acad Sci USA 98:9995–10000
Plaza S, Tearall KL, Zhao FJ, Buchner P, McGrath SP, Hawkesford MJ (2007) Expression and functional analysis of metal transporter genes in two contrasting ecotypes of the hyperaccumulator Thlaspi caerulescens. J Exp Bot 58:1717–1728
Rigola D, Fiers M, Vurro E, Aarts MGM (2006) The heavy metal hyperaccumulator Thlaspi caerulescens expresses many species-specific genes, as identified by comparative expressed sequence tag analysis. New Phytol 170:753–766
Tocquin P, Corbesier L, Havelange A, Pieltain A, Kurtem E, Bernier G, Perilleux C (2003) A novel high efficiency, low maintenance, hydroponic system for synchronous growth and flowering of Arabidopsis thaliana. BMC Plant Biology 3:2
van de Mortel JE, Villanueva LA, Schat H, Kwekkeboom CS, Moerland PD, Loren V, van Themaat E, Koornneef M, Aarts MGM (2006) Large expression difference in genes for iron and zinc homeostasis, stress response, and lignin biosynthesis distinguish roots of Arabidopsis thaliana and the related metal hyperaccumulator Thlaspi caerulescens. Plant Physiol 142:1127–1147
van de Mortel JE, Schat H, Moerland PD, Loren V, van Themaat E, van der Ent S, Blankestijn H, Ghandilyan A, Tsiatsiani S, Aarts MGM (2008) Expression differences for genes involved in lignin, glutathione and sulphate metabolism in response to cadmium in Arabidopsis thaliana and the related Zn/Cd-hyperaccumulator Thlaspi caerulescens. Plant Cell Environ 31:301–324
Varotto C, Maiwald D, Pesaresi P, Jahns P, Salamini F, Leister D (2002) The metal ion transporter IRT1 is necessary for iron homeostasis and efficient photosynthesis in Arabidopsis thaliana. Plant J 31:589–599
Vert G, Briat JF, Curie C (2001) The Arabidopsis IRT2 gene encodes a root-periphery iron transporter. Plant J 26:181–189
Vert G, Grotz N, Dedaldechamp F, Gaymard F, Guerinot ML, Briat JF, Curie C (2002) IRT1, an Arabidopsis transporter essential for iron uptake from the soil and for plant growth. Plant Cell 14:1223–1233
Weber M, Harada E, Vess C, Roepenack-Lahaye E, Clemens S (2004) Comparative microarray analysis of Arabidopsis thaliana and Arabidopsis halleri roots identifies nicotianamine synthase, a ZIP transporter and other genes as potential metal hyperaccumulation factors. Plant Journal 37:269–281
Acknowledgements
The authors thank Andrea Pirondini and Andrea Sacchani for kindly provide the T. caerulescens Monte Prinzera accession seeds, Bettine Aigner for the T. caerulescens Hochobir accession seeds and for T. minimum seeds, Paula Pongrac for the T. praecox seeds and Takafumi Mizuno for T. japonicum seeds, Judith van de Mortel for her support in preparing T. caerulescens cDNAs and Maarten Koornneef for critically reading the manuscript. The work was financially supported by the Interdisciplinary Research and Education Fund (INREF) of Wageningen University.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Author information
Authors and Affiliations
Corresponding author
Additional information
Responsible Editor: Jian Feng Ma.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Supplemental Fig. 1
Sequence alignment of predicted protein sequences of TcZNT5-LC from T. caerulescens and those of two additional T. caerulescens alleles (GenBank accessions AJ937739 (GA) and AF292029 (PR) and of AtZIP5 from A. thaliana (At1g05300, GenBank accession AC000098). Sequences are aligned using the ClustalW method. Identical residues of TcZNT5 and AtZIP5 are shaded. The putative transmembrane domains (TM) are numbered and overlined. A conserved stretch of repeated histidine residues between TM3 and TM4 and a conserved histidine residue in TM4 are highlighted in bold. (DOC 31 kb)
Supplemental Fig. 2
Sequence alignment of predicted protein sequences of TcZNT6-LC from T. caerulescens and of AtZIP6 from A. thaliana (At2g30080, GenBank accession AC004680). Sequences are aligned using ClustalW. Identical residues are shaded. The putative transmembrane domains (TM) are numbered and overlined. A conserved stretch of repeated histidine residues between TM3 and TM4, and a conserved histidine residue in TM4 are highlighted in bold. (DOC 28 kb)
Rights and permissions
Open Access This is an open access article distributed under the terms of the Creative Commons Attribution Noncommercial License (https://creativecommons.org/licenses/by-nc/2.0), which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
About this article
Cite this article
Wu, J., Zhao, FJ., Ghandilyan, A. et al. Identification and functional analysis of two ZIP metal transporters of the hyperaccumulator Thlaspi caerulescens . Plant Soil 325, 79–95 (2009). https://doi.org/10.1007/s11104-009-0151-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11104-009-0151-6