Abstract
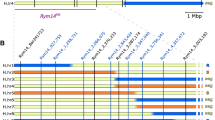
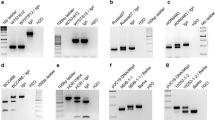
Barley yellow mosaic virus (BaYMV) disease seriously affects winter barley (Hordeum vulgare L.) production. Improvement of resistance to this disease can prevent yield losses. Based on previous reports, the gene rym7 (rmm7), conferring partial resistance to BaMMV, was assigned to chromosome 1H, closely linked to the centromere. In this study, newly developed barley genomic resources were applied to saturate the genetic map at the rym7 locus. Out of a set of 350 gene-based markers of chromosome 1H, we genetically assigned 23 to the rym7 region, delimiting the resistance locus to a 9.9-cM interval close to the centromere by genetic mapping in 53 doubled haploid progeny of a bi-parental mapping population. Nine gene-derived co-dominant markers co-segregated with the resistance locus. Among these, we identified the eukaryotic translation initiation factor Hv-eIF(iso)4E. Its homolog in Arabidopsis thaliana confers resistance to potyviruses. However, sequencing the entire open reading frame of resistant and susceptible genotypes could not reveal any sequence variation in exons of the gene. These results demonstrate how to combine newly developed barley genomic resources for rapid gene-based marker saturation. As a result, several easy-to-handle co-dominant markers have been identified for marker-assisted selection of rym7 in barley breeding.


Similar content being viewed by others
References
Adams MJ, Swaby AG, Jones P (1988) Confirmation of the transmission of Barley yellow mosaic-virus (BaYMV) by the fungus Polymyxa graminis. Ann Appl Biol 112(1):133–141
Boutin-Ganache I, Raposo M, Raymond M, Deschepper CF (2001) M13-tailed primers improve the readability and usability of microsatellite analyses performed with two different allele-sizing methods. Biotechniques 31(1):24–26
Chen JP, Adams MJ, Zhu FT, Wang ZQ, Chen J, Huang SZ, Zhang ZC (1996) Response of foreign barley cultivars to barley yellow mosaic virus at different sites in China. Plant Pathol 45(6):1117–1125
Chen J, Shi NN, Cheng Y, Diao A, Chen JP, Wilson TMA, Antoniw JF, Adams MJJ (1999) Molecular analysis of barley yellow mosaic virus isolates from China. Virus Res 64(1):13–21
Clark MF, Adams AN (1977) Characteristics of the microplate method of enzyme-linked immunosorbent assay for the detection of plant viruses. J Gen Virol 34(3):475–483
Comadran J, Kilian B, Russell J, Ramsay L, Stein N, Ganal M, Shaw P, Bayer M, Thomas W, Marshall D, Hedley P, Tondelli A, Pecchioni N, Francia E, Korzun V, Walther A, Waugh R (2012) Natural variation in a homolog of Antirrhinum CENTRORADIALIS contributed to spring growth habit and environmental adaptation in cultivated barley. Nat Genet 44(12):1388–1392
Doyle JJ, Doyle JL (1987) A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull 19:11–15
Duprat A, Caranta C, Revers F, Menand B, Browning KS, Robaglia C (2002) The Arabidopsis eukaryotic initiation factor (iso)4E is dispensable for plant growth but required for susceptibility to potyviruses. Plant J 32(6):927–934
Graner A, Streng S, Kellermann A, Proeseler G, Schiemann A, Peterka H, Ordon F (1999a) Molecular mapping of genes conferring resistance to soil-borne viruses in barley—an approach to promote understanding of host-pathogen interactions. Z Pflanzenk Pflanzen 106(4):405–410
Graner A, Streng S, Kellermann A, Schiemann A, Bauer E, Waugh R, Pellio B, Ordon F (1999b) Molecular mapping and genetic fine-structure of the rym5 locus encoding resistance to different strains of the Barley Yellow Mosaic Virus Complex. Theor Appl Genet 98(2):285–290
Habekuß A, Kuhne T, Kramer I, Rabenstein F, Ehrig F, Ruge-Wehling B, Huth W, Ordon F (2008) Identification of Barley mild mosaic virus isolates in Germany breaking rym5 resistance. J Phytopathol 156(1):36–41
Hariri D, Meyer M, Prud’homme H (2003) Characterization of a new barley mild mosaic virus pathotype in France. Eur J Plant Pathol 109(9):921–928
Huth W (1989) Ein Jahrzehnt Barley yellow mosaic virus in der Bundesrepublik Deutschland. Nachrichtenbl Dtsch Pflanzenschutzdienst 40:49–55
Huth W (1991) Verbreitung der Gelbmosaikviren BaYMV, BaMMV und BaYMV-2 und Screening von Gerstensorten auf Resistenz gegenüber BaYMV-2. Nachrichtenbl Dtsch Pflanzenschutzdienst 43:233–237
IBSC (2012) A physical, genetic and functional sequence assembly of the barley genome. Nature 491:711–716
International Brachypodium I (2010) Genome sequencing and analysis of the model grass Brachypodium distachyon. Nature 463(7282):763–768
Jenner CE, Nellist CF, Barker GC, Walsh JA (2010) Turnip mosaic virus (TuMV) is able to use alleles of both eIF4E and eIF(iso)4E from multiple loci of the diploid Brassica rapa. Mol Plant Microbe Interact 23(11):1498–1505
Kai H, Takata K, Tsukazaki M, Furusho M, Baba T (2012) Molecular mapping of Rym17, a dominant and rym18 a recessive barley yellow mosaic virus (BaYMV) resistance genes derived from Hordeum vulgare L. Theor Appl Genet 124(3):577–583
Kang BC, Yeam I, Jahn MM (2005) Genetics of plant virus resistance. Annu Rev Phytopathol 43:581–621
Kanyuka K, Ward E, Adams MJ (2003) Polymyxa graminis and the cereal viruses it transmits: a research challenge. Mol Plant Pathol 4(5):393–406
Kanyuka K, McGrann G, Alhudaib K, Hariri D, Adams MJ (2004) Biological and sequence analysis of a novel European isolate of Barley mild mosaic virus that overcomes the barley rym5 resistance gene. Arch Virol 149(8):1469–1480
Kanyuka K, Druka A, Caldwell DG, Tymon A, McCallum N, Waugh R, Adams MJ (2005) Evidence that the recessive bymovirus resistance locus rym4 in barley corresponds to the eukaryotic translation initiation factor 4E gene. Mol Plant Pathol 6(4):449–458
Kosambi DD (1944) The estimation of map distances from recombination values. Ann Eugen 12:172–175
Kühne T (2009) Soil-borne viruses affecting cereals—known for long but still a threat. Virus Res 141(2):174–183
Kühne T, Shi N, Proeseler G, Adams MJ, Kanyuka K (2003) The ability of a bymovirus to overcome the rym4-mediated resistance in barley correlates with a codon change in the VPg coding region on RNA1. J Gen Virol 84(Pt 10):2853–2859
Lellis AD, Kasschau KD, Whitham SA, Carrington JC (2002) Loss-of-susceptibility mutants of Arabidopsis thaliana reveal an essential role for eIF(iso)4E during Potyvirus infection. Curr Biol 12(12):1046–1051
Matsumoto T, Tanaka T, Sakai H, Amano N, Kanamori H, Kurita K, Kikuta A, Kamiya K, Yamamoto M, Ikawa H, Fujii N, Hori K, Itoh T, Sato K (2011) Comprehensive sequence analysis of 24,783 barley full-length cDNAs derived from 12 clone libraries. Plant Physiol 156(1):20–28
Mayer KF, Martis M, Hedley PE, Simkova H, Liu H, Morris JA, Steuernagel B, Taudien S, Roessner S, Gundlach H, Kubalakova M, Suchankova P, Murat F, Felder M, Nussbaumer T, Graner A, Salse J, Endo T, Sakai H, Tanaka T, Itoh T, Sato K, Platzer M, Matsumoto T, Scholz U, Dolezel J, Waugh R, Stein N (2011) Unlocking the barley genome by chromosomal and comparative genomics. Plant Cell 23(4):1249–1263
Neff MM, Turk E, Kalishman M (2002) Web-based primer design for single nucleotide polymorphism analysis. Trends Genet 18(12):613–615
Nishigawa H, Hagiwara T, Yumoto M, Sotome T, Kato T, Natsuaki T (2008) Molecular phylogenetic analysis of Barley yellow mosaic virus. Arch Virol 153(9):1783–1786
Nomura K, Kashiwazaki S, Hibino H, Inoue T, Nakata E, Tsuzaki Y, Okuyama S (1996) Biological and serological properties of strains of Barley mild mosaic virus. J Phytopathol 144(2):103–107
Ordon F, Ahlemeyer J, Werner K, Kohler W, Friedt W (2005) Molecular assessment of genetic diversity in winter barley and its use in breeding. Euphytica 146(1–2):21–28
Pellio B, Streng S, Bauer E, Stein N, Perovic D, Schiemann A, Friedt W, Ordon F, Graner A (2005) High-resolution mapping of the Rym4/Rym5 locus conferring resistance to the barley yellow mosaic virus complex (BaMMV, BaYMV, BaYMV-2) in barley (Hordeum vulgare ssp vulgare L.). Theor Appl Genet 110(2):283–293
Plumb RT, Lennon EA, Gutteridge RA (1986) The effects of infection by Barley yellow mosaic-virus on the yield and components of yield of barley. Plant Pathol 35(3):314–318
Robaglia C, Caranta C (2006) Translation initiation factors: a weak link in plant RNA virus infection. Trends Plant Sci 11(1):40–45
Ruffel S, Gallois JL, Moury B, Robaglia C, Palloix A, Caranta C (2006) Simultaneous mutations in translation initiation factors eIF4E and eIF(iso)4E are required to prevent pepper veinal mottle virus infection of pepper. J Gen Virol 87(Pt 7):2089–2098
Sato K, Nankaku N, Takeda K (2009) A high-density transcript linkage map of barley derived from a single population. Heredity 103(2):110–117
Schneeberger K, Ossowski S, Lanz C, Juul T, Petersen AH, Nielsen KL, Jorgensen J-E, Weigel D, Andersen SU (2009) SHOREmap: simultaneous mapping and mutation identification by deep sequencing. Nat Meth 6:550–551
Shahinnia F, Druka A, Frankowiak J, Morgante M, Waugh R, Stein N (2012) High resolution mapping of Dense spike-ar (dsp.ar) to the genetic centromere of barley chromosome 7H. Theor Appl Genet 124(2):373–384
Stein N, Perovic D, Kumlehn J, Pellio B, Stracke S, Streng S, Ordon F, Graner A (2005) The eukaryotic translation initiation factor 4E confers multiallelic recessive Bymovirus resistance in Hordeum vulgare (L.). Plant J 42(6):912–922
Stein N, Prasad M, Scholz U, Thiel T, Zhang HN, Wolf M, Kota R, Varshney RK, Perovic D, Grosse I, Graner A (2007) A 1,000-loci transcript map of the barley genome: new anchoring points for integrative grass genomics. Theor Appl Genet 114(5):823–839
Thiel T, Kota R, Grosse I, Stein N, Graner A (2004) SNP2CAPS: a SNP and INDEL analysis tool for CAPS marker development. Nucleic Acids Res 32(1):e5
Van Ooijen TW (2006) JoinMap 4, software for the calculation of genetic linkage maps in experimental populations. Kyazma BV, Wageningen, Netherlands
Varshney RK, Marcel TC, Ramsay L, Russell J, Roder MS, Stein N, Waugh R, Langridge P, Niks RE, Graner A (2007) A high density barley microsatellite consensus map with 775 SSR loci. Theor Appl Genet 114(6):1091–1103
Voorrips RE (2002) MapChart: software for the graphical presentation of linkage maps and QTLs. J Hered 93(1):77–78
Werner K, Friedt W, Ordon F (2005) Strategies for pyramiding resistance genes against the barley yellow mosaic virus complex (BaMMV, BaYMV, BaYMV-2). Mol Breed 16(1):45–55
Zheng T, Cheng Y, Chen JP, Antoniw JF, Adams MJ (1999) The occurrence of Barley mild mosaic virus (BaMMV) in China and the nucleotide sequence of its coat protein gene. J Phytopathol 147(4):229–234
Acknowledgments
We gratefully acknowledge Jelena Perovic (IPK) and Dörte Grau (JKI) for excellent technical assistance. The work was financed as part of the collaborative project “Plant KBBE II-ViReCrop” and was supported by a grant (FKZ.: 0315708) from the German Ministry of Education and Research (BMBF) to NS and FO.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Yang, P., Perovic, D., Habekuß, A. et al. Gene-based high-density mapping of the gene rym7 conferring resistance to Barley mild mosaic virus (BaMMV). Mol Breeding 32, 27–37 (2013). https://doi.org/10.1007/s11032-013-9842-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11032-013-9842-z