Abstract
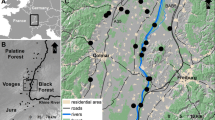
The European wildcat (Felis silvestris silvestris) is a focal species for conservation in many European countries. After a severe population decline during the 19th century, many populations became extinct or isolated. Within Germany, suitable wildcat habitat is assumed to be highly fragmented. We thus investigated fine-scale genetic structure of wildcat populations in Central Germany across two major potential barriers, the Rhine River with its valley and a major highway. We analyzed 260 hair and tissue samples collected between 2006 and 2011 in the Taunus and Hunsrück mountain ranges (3,500 km2 study area). We identified 188 individuals by genotyping 14 microsatellite loci, and found significant genetic substructure in the study area. Both the Rhine River and the highway were identified as significant barrier to gene flow. While the long-term effect of the river has led to stronger genetic differentiation in the river compared to the highway, estimates of current gene flow and relatedness across barriers indicated a similar or even stronger barrier effect to ongoing wildcat dispersal of the highway. Despite these barrier effects, we found evidence for the presence of recent migration across both the river and the highway. Our study thus suggests that although wildcats have the capability of dispersal across major anthropogenic and natural landscape barriers, these structures still lead to an effective isolation of populations as reflected by genetic analysis. The results strengthen the need for currently ongoing national strategies of wildcat conservation aiming for large scale habitat connectivity.





Similar content being viewed by others
References
Beerli P, Felsenstein J (1999) Maximum-likelihood estimation of migration rates and effective population numbers in two populations using a coalescent approach. Genetics 152(2):763–773
Beerli P, Felsenstein J (2001) Maximum likelihood estimation of a migration matrix and effective population sizes in n subpopulations by using a coalescent approach. Proc Natl Acad Sci USA 98(8):4563–4568. doi:10.1073/pnas.081068098
Birlenbach K, Klar N (2009) Aktionsplan für die Europäische Wildkatze in Deutschland. In: Fremuth W, Jedicke E, Kaphegyi T, Wachendörfer V, Weinzierl H (eds) Zukunft der Wildkatze in Deutschland. Ergebnisse des internationalen Wildkatzen-Symposiums 2008 in Wiesenfelden und Aktionsplan. Erich Schmidt Verlag, Berlin
Blanco JC, Cortes Y, Virgos E (2005) Wolf response to two kinds of barriers in an agricultural habitat in Spain. Can J Zool 83(2):312–323. doi:10.1139/z05-016
Broquet T, Ménard N, Petit E (2007) Noninvasive population genetics: a review of sample source, diet, fragment length and microsatellite motif effects on amplification success and genotyping error rates. Cons Genet 8(1):249–260. doi:10.1007/s10592-006-9146-5
Carmichael LE, Nagy JA, Larter NC, Strobeck C (2001) Prey specialization may influence patterns of gene flow in wolves of the Canadian Northwest. Mol Ecol 10(12):2787–2798. doi:10.1046/j.0962-1083.2001.01408.x
Cegelski CC, Waits LP, Anderson NJ (2003) Assessing population structure and gene flow in Montana wolverines (Gulo gulo) using assignment-based approaches. Mol Ecol 12(11):2907–2918. doi:10.1046/j.1365-294X.2003.01969.x
Chen C, Durand E, Forbes F, Francois O (2007) Bayesian clustering algorithms ascertaining spatial population structure: a new computer program and a comparison study. Mol Ecol Notes 7(5):747–756. doi:10.1111/j.1471-8286.2007.01769.x
Crawford NG (2010) SMOGD: software for the measurement of genetic diversity. Mol Ecol Res 10(3):556–557. doi:10.1111/j.1755-0998.2009.02801.x
Crosby MKA, Licht LE, Fu JZ (2009) The effect of habitat fragmentation on finescale population structure of wood frogs (Rana sylvatica). Cons Genet 10(6):1707–1718. doi:10.1007/s10592-008-9772-1
Cushman SA, Landguth EL (2010) Spurious correlations and inference in landscape genetics. Mol Ecol 19(17):3592–3602. doi:10.1111/j.1365-294X.2010.04656.x
Daniels MJ, Corbett L (2003) Redefining introgressed protected mammals: when is a wildcat a wild cat and a dingo a wild dog? Wildl Res 30(3):213–218
Devillard S, Jombart T, Pontier D (2009) Revealing cryptic genetic structuring in an urban population of stray cats (Felis silvestris catus). Mamm Biol 74(1):59–71. doi:10.1016/j.mambio.2008.01.001
Dickson BG, Jenness JS, Beier P (2005) Influence of vegetation, topography, and roads on cougar movement in southern California. J Wildl Manag 69(1):264–276
Driscoll CA, Menotti-Raymond M, Roca AL, Hupe K, Johnson WE, Geffen E, Harley EH, Delibes M, Pontier D, Kitchener AC, Yamaguchi N, O’Brien SJ, Macdonald DW (2007) The near eastern origin of cat domestication. Science 317(5837):519–523. doi:10.1126/science.1139518
Earl DA, vonHoldt BM (2011) STRUCTURE HARVESTER: a website and program for visualizing STRUCTURE output and implementing the Evanno method. Cons Gen Res doi:10.1007/s12686-011-9548-7In press
Eckert I, Suchentrunk F, Markov G, Hartl GB (2010) Genetic diversity and integrity of German wildcat (Felis silvestris) populations as revealed by microsatellites, allozymes, and mitochondrial DNA sequences. Mamm Biol 75(2):160–174. doi:10.1016/j.mambio.2009.07.005
Enserink M, Vogel G (2006) The carnivore comeback. Science 314(5800):746–749
Evanno G, Regnaut S, Goudet J (2005) Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Mol Ecol 14(8):2611–2620. doi:10.1111/j.1365-294X.2005.02553.x
Evett I, Weir B (eds) (1998) Interpreting DNA evidence: Statistical Genetics for Forensic Scientists. Sinauer Associates, Sunderland
Frantz AC, Cellina S, Krier A, Schley L, Burke T (2009) Using spatial Bayesian methods to determine the genetic structure of a continuously distributed population: clusters or isolation by distance? J Appl Ecol 46(2):493–505. doi:10.1111/j.1365-2664.2008.01606.x
Frantz AC, Pope LC, Etherington TR, Wilson GJ, Burke T (2010) Using isolation-by-distance-based approaches to assess the barrier effect of linear landscape elements on badger (Meles meles) dispersal. Mol Ecol 19(8):1663–1674. doi:10.1111/j.1365-294X.2010.04605.x
Frantz AC, Bertouille S, Eloy MC, Licoppe A, Chaumont F, Flamand MC (2012) Comparative landscape genetic analyses show a Belgian motorway to be a gene flow barrier for red deer (Cervus elaphus), but not wild boars (Sus scrofa). Mol Ecol 21(14):3445–3457. doi:10.1111/j.1365-294X.2012.05623.x
Gauffre B, Estoup A, Bretagnolle V, Cosson JF (2008) Spatial genetic structure of a small rodent in a heterogeneous landscape. Mol Ecol 17(21):4619–4629. doi:10.1111/j.1365-294X.2008.03950.x
Götz M, Jerosch S (2010) Wildkatzen und Straßen—Ermittlung von Unfallschwerpunkten im Ostharz. Natuschutz im Land Sachsen-Anhalt 47:26–33
Goudet J (1995) FSTAT (Version 1.2): a computer program to calculate F-statistics. J Hered 86(6):485–486
Guillot G, Leblois R, Coulon A, Frantz AC (2009) Statistical methods in spatial genetics. Mol Ecol 18(23):4734–4756. doi:10.1111/j.1365-294X.2009.04410.x
Hardy OJ, Vekemans X (2002) SPAGeDI: a versatile computer program to analyse spatial genetic structure at the individual or population levels. Mol Ecol Notes 2(4):618–620. doi:10.1046/j.1471-8286.2002.00305.x
Heller R, Siegismund HR (2009) Relationship between three measures of genetic differentiation G ST, D EST and G ST ’: how wrong have we been? Mol Ecol 18(10):2080–2083. doi:10.1111/j.1365-294X.2009.04185.x
Herrmann M, Vogel C (2005) Wildkatze Felis silvestris silvestris Schreber, 1777. In: Braun M, Dieterlen F (eds) Die Säugetiere Baden-Württembergs, vol 2. Ulmer, Stuttgart, pp 363–376
Hertwig ST, Schweizer M, Stepanow S, Jungnickel A, Bohle UR, Fischer MS (2009) Regionally high rates of hybridization and introgression in German wildcat populations (Felis silvestris, Carnivora, Felidae). J Zool Syst Evol Res 47(3):283–297. doi:10.1111/j.1439-0469.2009.00536.x
Huck M, Jedrzejewski W, Borowik T, Miłosz-Cielma M, Schmidt K, Jedrzejewska B, Nowak S, Mysłajek RW (2010) Habitat suitability, corridors and dispersal barriers for large carnivores in Poland. Acta Theriol 55(2):177–192. doi:10.4098/j.at.0001-7051.114.2009
Jakobsson M, Rosenberg NA (2007) CLUMPP: a cluster matching and permutation program for dealing with label switching and multimodality in analysis of population structure. Bioinformatics 23(14):1801–1806. doi:10.1093/bioinformatics/btm233
Janecka JE, Jackson R, Yuquang Z, Diqiang L, Munkhtsog B, Buckley-Beason V, Murphy WJ (2008) Population monitoring of snow leopards using noninvasive collection of scat samples: a pilot study. Anim Conserv 11(5):401–411. doi:10.1111/j.1469-1795.2008.00195.x
Jost L (2008) G ST and its relatives do not measure differentiation. Mol Ecol 17(18):4015–4026. doi:10.1111/j.1365-294X.2008.03887.x
Kaczensky P, Knauer F, Krze B, Jonozovic M, Adamic M, Gossow H (2003) The impact of high speed, high volume traffic axes on brown bears in Slovenia. Biol Conserv 111(2):191–204. doi:10.1016/S0006-3207(02)00273-2
Kalinowski ST, Taper ML, Marshall TC (2007) Revising how the computer program CERVUS accommodates genotyping error increases success in paternity assignment. Mol Ecol 16(5):1099–1106. doi:10.1111/j.1365-294X.2007.03089.x
Kéry M, Gardner B, Stoeckle T, Weber D, Royle JA (2011) Use of spatial capture-recapture modeling and DNA data to estimate densities of elusive animals. Conserv Biol 25(2):356–364. doi:10.1111/j.1523-1739.2010.01616.x
Klar N (2007) Habitatwahl in der Südeifel. Die Wildkatze in der Eifel—Habitate, Ressourcen, Streifgebiete. Laurenti-Verlag, Bielefeld
Klar N, Fernández N, Kramer-Schadt S, Herrmann M, Trinzen M, Büttner I, Niemitz C (2008) Habitat selection models for European wildcat conservation. Biol Conserv 141(1):308–319. doi:10.1016/j.biocon.2007.10.004
Klar N, Herrmann M, Kramer-Schadt S (2009) Effects and mitigation of road impacts on individual movement behavior of wildcats. J Wildl Manag 73(5):631–638. doi:10.2193/2007-574
Klar N, Herrmann M, Henning-Hahn M, Pott-Dörfer B, Hofer H, Kramer-Schadt S (2012) Between ecological theory and planning practice: (re-)connecting forest patches for the wildcat in Lower Saxony, Germany. Landsc Urban Plan 105(4):376–384. doi:10.1016/j.landurbplan.2012.01.007
Krüger M, Hertwig ST, Jetschke G, Fischer MS (2009) Evaluation of anatomical characters and the question of hybridization with domestic cats in the wildcat population of Thuringia, Germany. J Zool Syst Evol Res 47(3):268–282
Landguth EL, Cushman SA, Schwartz MK, McKelvey KS, Murphy M, Luikart G (2010) Quantifying the lag time to detect barriers in landscape genetics. Mol Ecol 19(19):4179–4191. doi:10.1111/j.1365-294X.2010.04808.x
Lecis R, Pierpaoli M, Biro ZS, Szemethy L, Ragni B, Vercillo F, Randi E (2006) Bayesian analyses of admixture in wild and domestic cats (Felis silvestris) using linked microsatellite loci. Mol Ecol 15(1):119–131
Legendre P, Dale MRT, Fortin MJ, Gurevitch J, Hohn M, Myers D (2002) The consequences of spatial structure for the design and analysis of ecological field surveys. Ecography 25(5):601–615
Leng L, Zhang DX (2011) Measuring population differentiation using G ST or D? A simulation study with microsatellite DNA markers under a finite island model and nonequilibrium conditions. Mol Ecol 20(12):2494–2509. doi:10.1111/j.1365-294X.2011.05108.x
Loiselle BA, Sork VL, Nason J, Graham C (1995) Spatial genetic structure of a tropical understory shrub, Psychotria officinalis (Rubiaceae). Am J Bot 82(11):1420–1425
Marsh DM, Page RB, Hanlon TJ, Corritone R, Little EC, Seifert DE, Cabe PR (2008) Effects of roads on patterns of genetic differentiation in red-backed salamanders Plethodon cinereus. Cons Genet 9(3):603–613. doi:10.1007/s10592-007-9377-0
Menotti-Raymond M, David VA, Lyons LA, Schaffer AA, Tomlin JF, Hutton MK, O’Brien SJ (1999) A genetic linkage map of microsatellites in the domestic cat (Felis catus). Genomics 57(1):9–23. doi:10.1006/geno.1999.5743
Millions DG, Swanson BJ (2007) Impact of natural and artificial barriers to dispersal on the population structure of bobcats. J Wildl Manag 71(1):96–102. doi:10.2193/2005-563
Mills LS, Allendorf FW (1996) The one-migrant-per-generation rule in conservation and management. Conserv Biol 10(6):1509–1518. doi:10.1046/j.1523-1739.1996.10061509.x
Mills LS, Citta JJ, Lair KP, Schwartz MK, Tallmon DA (2000) Estimating animal abundance using noninvasive DNA sampling: promise and pitfalls. Ecol Appl 10:283–294. doi:10.1890/1051-0761(2000)010[0283:EAAUND]2.0.CO;2
Mölich T, Klaus S, Nöllert A (2003) Die Wildkatze in Thüringen. Landschaftspflege und Naturschutz in Thüringen 40(4) Sonderheft
Oliveira R, Godinho R, Randi E, Ferrand N, Alves PC (2007) Molecular analysis of hybridisation between wild and domestic cats (Felis silvestris) in Portugal: implications for conservation. Cons Genet 9(1):1–11
Palsbøll PJ, Peery ZM, Bérubé M (2010) Detecting populations in the ‘ambiguous’ zone: kinship-based estimation of population structure at low genetic divergence. Mol Ecol Res 10(5):797–805. doi:10.1111/j.1755-0998.2010.02887.x
Peakall R, Smouse PE (2006) GENALEX 6: genetic analysis in Excel. Population genetic software for teaching and research. Mol Ecol Notes 6(1):288–295. doi:10.1111/j.1471-8286.2005.01155.x
Piechocki R (1990) Die Wildkatze. Die Neue Brehm-Bücherei. Westarp Wissenschaften, Lutherstadt
Pierpaoli M, Biro ZS, Herrmann M, Hupe K, Fernandes M, Ragni B, Szemethy L, Randi E (2003) Genetic distinction of wildcat (Felis silvestris) populations in Europe, and hybridization with domestic cats in Hungary. Mol Ecol 12(10):2585–2598. doi:10.1046/j.1365-294X.2003.01939.x
Pilgrim KL, McKelvey KS, Riddle AE, Schwartz MK (2005) Felid sex identification based on noninvasive genetic samples. Mol Ecol Notes 5(1):60–61. doi:10.1111/j.1471-8286.2004.00831.x
Pritchard JK, Stephens M, Donnelly P (2000) Inference of population structure using multilocus genotype data. Genetics 155:945–959
R Development Core Team. R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna. http://www.R-project.org (2009)
Raimer F (1988) Die Wildkatze in Hessen und Niedersachsen. Biotop, Umwelt, Verbreitung, Bestandsentwicklung, Gefährdung, Schutz, Projektarbeit an der Gesamthochschule Kassel. Witzenhausen
Raimer F (1989) Die Wildkatze in Hessen und Niedersachsen—Fortführung der Projektarbeit von 1988, Historischer Überblick, Biologie. Meinungsbild, Kassel
Belkhir K, Borsa P, Chikhi L, Raufaste N, F. B (1996–2004) GENETIX 4.05, logiciel sous Windows TM pour la génétique des populations. Laboratoire Génome, Populations, Interactions, CNRS UMR 5000, Université de Montpellier II, Montpellier (France)
Raymond M, Rousset F (1995) Genepop (Version-1.2): population-genetics software for exact tests and ecumenicism. J Hered 86(3):248–249
Riley SPD, Pollinger JP, Sauvajot RM, York EC, Bromley C, Fuller TK, Wayne RK (2006) A southern California freeway is a physical and social barrier to gene flow in carnivores. Mol Ecol 15(7):1733–1741. doi:10.1111/j.1365-294X.2006.02907.x
Rosenberg NA (2004) DISTRUCT: a program for the graphical display of population structure. Mol Ecol Notes 4(1):137–138. doi:10.1046/j.1471-8286.2003.00566.x
Say L, Devillard S, Léger F, Pontier D, Ruette S (2012) Distribution and spatial genetic structure of European wildcat in France. Anim Cons 15(1):18–27. doi:10.1111/j.1469-1795.2011.00478.x
Schwartz MK, McKelvey KS (2009) Why sampling scheme matters: the effect of sampling scheme on landscape genetic results. Conserv Genet 10(2):441–452. doi:10.1007/s10592-008-9622-1
Simon O (2007) Wildkatzen-Wegeplan Hessen—Biotopverbundkonzept für die Wildkatze Felis silvestris silvestris in Hessen im Rahmen des BUND-Projektes Ein Rettungsnetz für die Wildkatze. BUND Hessen, Frankfurt
Steyer K (2009) Genetisches Monitoring der Wildkatze, Felis silvestris, in Deutschland. Frankfurt am Main
Steyer K, Simon O, Kraus RHS, Haase P, Nowak C (2013) Hair trapping with valerian-treated lure sticks as a tool for genetic wildcat monitoring in low-density habitats. Eur J Wildl Res 59(1):39–46. doi:10.1007/s10344-012-0644-0
Taberlet P, Waits LP, Luikart G (1999) Noninvasive genetic sampling: look before you leap. Trends Ecol Evol 14(8):323–327. doi:10.1016/S0169-5347(99)01637-7
Turok I, Mykhnenko V (2007) The trajectories of European cities, 1960–2005. Cities 24(3):165–182. doi:10.1016/j.cities.2007.01.007
Valière N (2002) GIMLET: a computer program for analysing genetic individual identification data. Mol Ecol Notes 10:1046. doi:10.1046/j.1471-8286.2002.00228.x-i2
Waits LP, Paetkau D (2005) Noninvasive genetic sampling tools for wildlife biologists: a review of applications and recommendations for accurate data collection. J Wildl Manag 69(4):1419–1433. doi:10.2193/0022-541X(2005)69[1419:NGSTFW]2.0.CO;2
Whitlock MC (2011) G ST ′ and D do not replace F ST. Mol Ecol 20(6):1083–1091. doi:10.1111/j.1365-294X.2010.04996.x
Wright S (1951) The genetical structure of populations. Ann Eugen 15:323–354. doi:10.1111/j.1469-1809.1949.tb02451.x
Acknowledgments
We are grateful to numerous persons who helped to conduct this research in the federal states of Hesse and Rhineland-Palatine, in particular to Martina Denk for data collection and the Hessen-Forst-Forsteinrichtung und Naturschutz (FENA) for kind permission of data use in the Rheingau-Taunus region. Olaf Simon (Institut für Tierökologie und Naturbildung), Thomas Mölich and Burkhard Vogel (Bund für Umwelt und Naturschutz, BUND), and Peter Haase (Senckenberg Research Institute and Natural History Museum) provided valuable help and discussion. We are particularly grateful to Alain Frantz for providing help in analysis and data interpretation, and to Peter Beerli for supporting us in making good choices for settings in MIGRATE-n. Kyle Tomlinson helped revising the English of the present paper, and extensive comments of Seth Riley, two anonymous reviewers, and the guest editor Niko Balkenhol improved the quality of paper. Further thanks go to Paul Peitz, Dietrich Nord and Doris Warlich for their interest in the project and patrolling lure sticks. We thank K. Volmer, W. Hecht, S. Steeb (University of Gießen), for providing coordinates. MIGRATE-n calculations carried out in this manuscript were performed on the CSC HPC cluster FUCHS of the J. W. Goethe University Frankfurt, Germany. RHSK was funded by grant SAW-2011-SGN-3 from the Leibniz Association (Germany).
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Hartmann, S.A., Steyer, K., Kraus, R.H.S. et al. Potential barriers to gene flow in the endangered European wildcat (Felis silvestris). Conserv Genet 14, 413–426 (2013). https://doi.org/10.1007/s10592-013-0468-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10592-013-0468-9