Abstract
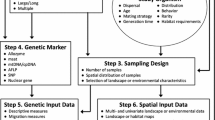
There has been a recent trend in genetic studies of wild populations where researchers have changed their sampling schemes from sampling pre-defined populations to sampling individuals uniformly across landscapes. This reflects the fact that many species under study are continuously distributed rather than clumped into obvious “populations”. Once individual samples are collected, many landscape genetic studies use clustering algorithms and multilocus genetic data to group samples into subpopulations. After clusters are derived, landscape features that may be acting as barriers are examined and described. In theory, if populations were evenly sampled, this course of action should reliably identify population structure. However, genetic gradients and irregularly collected samples may impact the composition and location of clusters. We built genetic models where individual genotypes were either randomly distributed across a landscape or contained gradients created by neighbor mating for multiple generations. We investigated the influence of six different sampling protocols on population clustering using program STRUCTURE, the most commonly used model-based clustering method for multilocus genotype data. For models where individuals (and their alleles) were randomly distributed across a landscape, STRUCTURE correctly predicted that only one population was being sampled. However, when gradients created by neighbor mating existed, STRUCTURE detected multiple, but different numbers of clusters, depending on sampling protocols. We recommend testing for fine scale autocorrelation patterns prior to sample clustering, as the scale of the autocorrelation appears to influence the results. Further, we recommend that researchers pay attention to the impacts that sampling may have on subsequent population and landscape genetic results.






Similar content being viewed by others
References
Barbujani G (1987) Autocorrelation of gene frequencies under isolation by distance. Genetics 117:777–782
Barbujani G, Sokal R (1989) Zones of sharp genetic change in Europe are also linguistic boundaries. Proc Natl Acad Sci USA 87:1816–1819
Bertorelle G, Barbujani G (1995) Analysis of DNA diversity by spatial autocorrelation. Genetics 140:811–819
Burton C, Krebs CJ, Taylor EB (2002) Population genetic structure of the cyclic snowshoe hare (Lepus americanus) in southwestern Yukon, Canada. Mol Ecol 11:1689–1701
Cegelski C, Waits L, Anderson N (2003) Assessing population substructure and gene flow in Montana wolverines (Gulo gulo) using assignment-based approaches. Mol Ecol 12:2907–2918
Cegelski CC, Waits LP, Anderson NJ et al (2006) Genetic diversity and population structure of wolverine (Gulo gulo) populations at the southern edge of their current distribution in North America with implications for genetic viability. Conserv Genet 7:1566–1572
Chen C, Durand E, Forbes F, François O (2007) Bayesian clustering algorithms ascertaining spatial population structure: a new computer program and a comparison study. Mol Ecol Notes 7:747–756
Corander J, Waldmann P, Marttinen P, Sillanpaa MJ (2004) BAPS 2: enhanced possibilities for the analysis of genetic population structure. Bioinformatics 20:2363–2369
Cushman SA, McKelvey KS, Hayden J, Schwartz MK (2006) Gene-flow in complex landscapes: testing multiple hypotheses with causal modeling. Am Nat 168:486–499
Dawson K, Belkhir K (2001) A Bayesian approach to the identification of panmictic populations and the assignment of individuals. Genet Res 78:59–77
Double MC, Peakall R, Beck NR, Cockburn A (2005) Dispersal, philopatry, and infidelity: dissecting local genetic structure in superb fairly-wrens (Malurus cyaneus). Evolution 59:625–635
Epperson BK, Li T (1996) Measurement of genetic structure within population using Moran’s spatial autocorrelation statistics. Proc Natl Acad Sci USA 93:10528–10532
Evanno G, Regnaut S, Goudet J (2005) Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Mol Ecol 14:2611–2620
Falush D, Stephens M, Pritchard JK (2003) Inference of population structure using multilocus genotype data: linked loci and correlated allele frequencies. Genetics 164:1567–1587
Falush D, Stephens M, Pritchard JK (2007) Inference of population structure using multilocus genotype data: dominant markers and null alleles. Mol Ecol Notes 7:574–578
Fonseca DM, Keyghobadi N, Malcolm CA et al (2004) Emerging vectors in the Culex pipiens complex. Science 303:1535–1538
François O, Ancelet S, Guillot G (2006) Bayesian clustering using hidden Markov random fields in spatial population genetics. Genetics 174:805–816
Funk WC, Blouin MS, Corn PS et al (2005) Population structure of Columbia spotted frogs (Rana luteiventris) is strongly affected by the landscape. Mol Ecol 14:483–496
Gamache I, Jaramillo-Correa JP, Payette S, Bousquet J (2003) Diverging patters of mitochondrial and nuclear DNA diversity in subarctic black spruce: imprint of a founder effect associated with postglacial colonization. Mol Ecol 12:891–901
Goudet J, (1995) Fstat version 1.2: a computer program to calculate Fstatistics. Journal of Heredity, 86: 485–486.
Guillot G, Mortier F, Estoup A (2005a) GENELAND: a computer package for landscape genetics. Mol Ecol Notes 5:712–715
Guillot G, Estoup A, Mortier F, Cosson J (2005b) A spatial statistical model for landscape genetics. Genetics 170:1261–1280
Hicks JF, Rachlow JL, Rhodes OE, Williams CL, Waits LP (2007) Reintroduction and genetic structure: rocky Mountain elk in Yellowstone and the western states. J Mamm 88:129–138
Holderegger R, Wagner HH (2006) A brief guide to landscape genetics. Landsc Ecol 21:793–796
Jorde PE, Schweder T, Bickham JW et al (2007) Detecting genetic structure in migrating bowhead whales off the coast of Barrow, Alaska. Mol Ecol 16:1993–2004
Kalinowski ST, Taper ML, Marshall TC (2007) Revising how the computer program CERVUS accommodates genotyping error increases success in paternity assignment. Mol Ecol 16:1099–1106
Kimura M, Weiss GH (1964) The stepping stone model of population structure and the decrease of genetic correlation with distance. Genetics 49:561–576
Kyle CJ, Strobeck C (2002) Connectivity of peripheral and core populations of North American wolverines. J Mamm 83:1141–1150
Latch E, Rhodes OE Jr (2006) Evidence for bias in estimates of local genetic structure due to sampling scheme. Anim Conserv 9:308–315
Latch E, Dharmarajan G, Glaubitz JC, Rhodes OE Jr (2006) Relative performance of Bayesian clustering software for inferring population substructure and individual assignment at low levels of population differentiation. Conserv Genet 7:1566–1572
Legendre P, Dale MRT, Fortin M-J, Gurevitch J, Hohn M, Myers D (2002) The consequences of spatial structure for the design and analysis of ecological field surveys. Ecography 25:601–616
Legendre P, Dale MRT, Fortin M-J, Casgrain P, Gurevitch J (2004) Effects of spatial structures on the results of field experiments. Ecology 85:3202–3214
Malecot G (1973) Isolation by distance. In: Morton NE (ed) Genetic structure of populations. University of Hawaii Press, Honolulu, pp 72–75
Manel S, Schwartz MK, Luikart G, Taberlet P (2003) Landscape genetics: the combination of landscape ecology and population genetics. Trends Ecol Evol 18:189–197
Millions DG, Swanson BJ (2007) Impact of natural and artificial barriers to dispersal of the population structure of bobcats. J Wildl Manage 71:96–102
Morton NE (1973) Genetic structure of populations. University of Hawaii Press, Honolulu
Mossman CA, Waser PM (2001) Effects of habitat fragmentation on population genetic structure in the white-footed mouse (Peromyscus leucopus). Can J Zool 79:285–295
Musiani M, Leonard JA, Cluff HD et al (2007) Differentiation of tundra/taiga and boreal coniferous forest wolves: genetics, coat color and association with migratory caribou. Mol Ecol 16:4149–4170
Natoli A, Birkun A, Aguilar A, Lopez A, Hoelzel AR (2005) Habitat structure and the dispersal of male and female bottlenose dolphins (Tursiops truncatus). Proc R Soc Lond B 272:1217–1226
Pardini AT, Jones CS, Noble LR et al (2001) Sex-biased dispersal of great white sharks. Science 412:139–140
Parra FC, Amado RC, Lambertucci JR et al (2003) Color and genomic ancestry in Brazilians. Proc Natl Acad Sci USA 100:177–182
Peakall R, Smouse PE (2006) GENALEX 6: genetic analysis in Excel. Population genetic software for teaching and research. Mol Ecol Notes 6:288–295
Piertney SB, MacColl ADC, Bacon PJ, Dallas JF (1998) Local genetic structure in red grouse (Lagopus lagopus scoticus): evidence from microsatellite DNA markers. Mol Ecol 7:1645–1654
Pritchard JK, Stephens M, Donnelly P (2000) Inference of population structure using multilocus genotype data. Genetics 155:945–959
Pritchard JK, Wen X, Falush D (2007) Documentation for structure software: version 2.2. University of Chicago, Chicago, pp 1–36
Riley SPD, Pollinger JP, Sauvajot RM et al (2006) A southern California freeway is a physical and social barrier to gene flow in carnivores. Mol Ecol 15:1733–1741
Repaci V, Stow AJ, Briscoe DA (2006) Fine-scale genetic structure, co-founding and multiple mating in the Australian allodapine bee (Exoneura robusta). J Zool 270:687–691
Rosenberg NA, Pritchard JK, Weber JL et al (2002) Genetic structure of human populations. Science 298:2381–2385
Rosenberg NA, Li LM, Ward R, Pritchard JK (2003) Informativeness of genetic markers for inference of ancestry. Am J Hum Genet 73:1402–1422
Rosenberg NA, Mahajan S, Ramachandran S et al (2005) Clines, clusters, and the effect of study design on the inference of human population structure. PLoS Genetics 1:660–671
Schwartz MK, Mills LS, Ortega YK, Ruggiero LF, Allendorf FW (2003) Landscape location affects genetic variation of Canada lynx (Lynx canadensis). Mol Ecol 12:1807–1816
Schwartz MK, Pilgrim KL, McKelvey KS et al (2004) Hybridization between Canada lynx and bobcats: genetic results and management implications. Conservation Genetics 5:349–355
Schwartz MK, Cushman SA, McKelvey KS, Hayden J, Engkjer C (2006) Detecting genotyping errors and describing American black bear movement in northern Idaho. Ursus 17:138–148
Serre D, Paabo S (2004) Evidence for gradients of human genetic diversity within and among continents. Genomics Res 14:1679–1685
Sokal RR, Oden NL (1978a) Spatial autocorrelation in biology, 2. Some biological implications and four applications of evolutionary and ecological interest. Biol J Linn Soc 10:229–249
Sokal RR, Oden NL (1978b) Spatial autocorrelation in biology. 1. Methodology. Biol J Linn Soc 10:199–228
Sokal RR, Oden NL, Thomson BA (1998) Local spatial autocorrelation in biological variables. Biol J Linn Soc 65:41–62
Storfer A, Murphy MA, Evans JS et al (2007) Putting the ‘landscape’ in landscape genetics. Heredity 98:128–142
Tallmon DA, Draheim HM, Mills LS, Allendorf FW (2002) Insights into recently fragmented vole populations from combined genetic and demographic data. Mol Ecol 11:699–709
Tero N, Aspi J, Siikamaki P, Jakalaniemi A, Tuomi J (2003) Genetic structure and gene flow in a metapopulation of an endangered plant species, Silene tatarica. Mol Ecol 12:2073–2085
Witherspoon DJ et al (2006) Human population genetic structure and diversity inferred from polymorphic L1 (LINE-1) and Alu insertions. Hum Hered 62:30–46
Acknowledgements
This research was supported by a Presidential Early Career Award for Science and Engineering to MKS. We thank Sam Cushman, Gordon Luikart, Fred Allendorf, and the Allendorf lab seminar group for thoughts on this work. In addition, we thank Jodi Copeland for help running simulations. We thank Jonathan Pritchard, Eric Anderson, and two anonymous reviewers for comments on earlier drafts of this manuscript.
Author information
Authors and Affiliations
Corresponding author
Additional information
The U.S. Government's right to retain a non-exclusive, royalty-free license in and to any copyright is acknowledged.
Rights and permissions
About this article
Cite this article
Schwartz, M.K., McKelvey, K.S. Why sampling scheme matters: the effect of sampling scheme on landscape genetic results. Conserv Genet 10, 441–452 (2009). https://doi.org/10.1007/s10592-008-9622-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10592-008-9622-1