Abstract
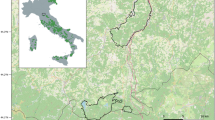
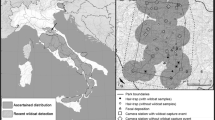
Wildcats are among the most elusive and least investigated carnivores in Central Europe. Here, we propose a hair-trapping method that allows reliable detection of wildcat presence even in low-density habitats. The trap is simple, consisting of a wooden stick with valerian as cat attractant. We performed non-invasive genetic wildcat monitoring in the Kellerwald-Edersee National Park, Germany, between 2007 and 2011. Our results provide the first evidence of wildcat presence in this region. Microsatellite analysis and mtDNA sequencing of hair samples furthermore confirm the existence of at least six individuals (males and females) in the study region. Four individuals were detected over two consecutive years, suggesting the resident status of wildcats in this area. Our results show that the lure stick method releases its full potential when combined with genetic analysis and is a sensitive tool which not only enables the detection of wildcat presence but also provides individual identification, even in recently colonised low-density areas.



Similar content being viewed by others
References
Anile S, Bizzarri L, Ragni B (2010) Estimation of European wildcat population size in Sicily (Italy) using camera trapping and capture-recapture analyses. Ital J Zool 77:241–246
Beja-Pereira A, Oliveira R, Alves PC et al (2009) Advancing ecological understandings through technological transformations in noninvasive genetics. Mol Ecol Resour 9:1279–1301
Bizzarri L, Lacrimini M, Ragni B (2010) Live capture and handling of the European wildcat in central Italy. Hystrix 21:73–82
Bonin A, Bellemain E, Eidesen PB et al (2004) How to track and assess genotyping errors in population genetics studies. Mol Ecol 13:3261–3273
Broquet T, Ménard N, Petit E (2007) Noninvasive population genetics: a review of sample source, diet, fragment length and microsatellite motif effects on amplification success and genotyping error rates. Conserv Genet 8:249–260
Daniels MJ, Beaumont MA, Johnson PJ et al (2001) Ecology and genetics of wild-living cats in the north-east of Scotland and the implications for the conservation of the wildcat. J Appl Ecol 38:146–161
Eckert I, Suchentrunk F, Markov G, Hartl GB (2009) Genetic diversity and integrity of German wildcat (Felis silvestris) populations as revealed by microsatellites, allozymes, and mitochondrial DNA sequences. Mamm Biol 75:160–174
Gagneux P, Boesch C, Woodruff DS (1997) Microsatellite scoring errors associated with noninvasive genotyping based on nuclear DNA amplified from shed hair. Mol Ecol 6:861–868
Goossens B, Waits LP, Taberlet P (1998) Plucked hair samples as a source of DNA: reliability of dinucleotide microsatellite genotyping. Mol Ecol 7:1237–1241
Hall TA (1999) BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucl Acids Symp Ser 41:95–98
Hertwig ST, Schweizer M, Stepanow S et al (2009) Regionally high rates of hybridization and introgression in German wildcat populations (Felis silvestris, Carnivora, Felidae). J Zool Syst Evol Res 47:283–297
Hupe K (2007) Untersuchung zum Vorkommen der Wildkatze (Felis silvestris silvestris) in Wäldern und bewaldeten Höhenzügen zwischen Solling und Hainberg im Hinblick auf eine mögliche Vernetzung der Harz- und Sollingpopulation. Inform.d. Naturschutz Niedersachs 27:38–45
Hupe K, Simon O (2007) Die Lockstockmethode - eine nicht invasive Methode zum Nachweis der Europäischen Wildkatze (Felis s. silvestris). Inform d Naturschutz Niedersachs 27:66–69
Jerosch S, Götz M, Klar N, Roth M (2010) Characteristics of diurnal resting sites of the endangered European wildcat (Felis silvestris silvestris): implications for its conservation. J Nat Conserv 18:45–54
Klar N, Fernández N, Kramer-Schadt S et al (2008) Habitat selection models for European wildcat conservation. Biol Conserv 141:308–319
Kocher TD, Thomas WK, Meyer A et al (1989) Dynamics of mitochondrial DNA evolution in animals: amplification and sequencing with conserved primers. P Natl Acad Sci USA 86:6196–6200
Krüger M, Hertwig ST, Jetschke G, Fischer MS (2009) Evaluation of anatomical characters and the question of hybridization with domestic cats in the wildcat population of Thuringia, Germany. J Zool Syst Evol Res 47:268–282
Lozano J, Virgos E, Malo AF et al (2003) Importance of scrub-pastureland mosaics for wild-living cats occurrence in a Mediterranean area: implications for the conservation of the wildcat (Felis silvestris). Biodivers Conserv 12:921–935
Menotti-Raymond M, David VA, Lyons LA et al (1999) A genetic linkage map of microsatellites in the domestic cat (Felis catus). Genomics 57:9–23
Monterroso P, Brito JC, Ferreras P, Alves PC (2009) Spatial ecology of the European wildcat in a Mediterranean ecosystem: dealing with small radio-tracking datasets in species conservation. J Zool 279:27–35
Monterroso P, Alves PC, Ferreras P (2011) Evaluation of attractants for non-invasive studies of Iberian carnivore communities. Wildl Res 38:446
Navidi W, Arnheim N, Waterman MS (1992) A multiple-tubes approach for accurate genotyping of very small DNA samples by using PCR: statistical considerations. Am J Hum Genet 50:347–359
Nowak C, Steyer K (2009) Abschlußbericht Genetisches Monitoring der Wildkatze im Rahmen des Projektes “Ein Rettungsnetz für die Wildkatze”- Teilbereich Kontrolle. Conservation Genetics Group, Senckenberg Gelnhausen
Oliveira R, Godinho R, Randi E et al (2007) Molecular analysis of hybridisation between wild and domestic cats (Felis silvestris) in Portugal: implications for conservation. Conserv Genet 9:1–11
Okarma H, Sniezko S, Olszanska A (2002) The occurrence of wildcat in the Polish Carpathian Mountains. Acta Theriol 47:499–504
O'Brien J, Devillard S, Say L et al (2009) Preserving genetic integrity in a hybridising world: are European Wildcats (Felis silvestris silvestris) in eastern France distinct from sympatric feral domestic cats? Biodivers Conserv 18:2351–2360
Peakall R, Smouse PE (2006) GENALEX 6: genetic analysis in Excel. Population genetic software for teaching and research. Mol Ecol Notes 6:288–295
Piechocki R (1990) Die Wildkatze. Die Neue Brehm-Bücherei, Wittenberg-Lutherstadt
Pierpaoli M, Biro ZS, Herrmann M et al (2003) Genetic distinction of wildcat (Felis silvestris) populations in Europe, and hybridization with domestic cats in Hungary. Mol Ecol 12:2585–2598
Pilgrim KL, McKelvey KS, Riddle AE, Schwartz MK (2005) Felid sex identification based on noninvasive genetic samples. Mol Ecol Notes 5:60–61
Pritchard JK, Stephensa M, Donnellya P (2000) Inference of population structure using multilocus genotype data. Genetics 155:945–959
Raimer F (1988) Die Wildkatze in Hessen und Niedersachsen. University of Kassel, Diploma thesis
Semrau M (2000) Studie zur Situation der Wildkatze im Kellerwald. University of Göttingen, Diploma thesis
Potocnik H, Kljun F, Racnik J et al (2002) Experience obtained from box trapping and handling wildcats in Slovenia. Acta Theriol 47:211–219
Ruell EW, Crooks KR (2007) Evaluation of noninvasive genetic sampling methods for felid and canid populations. J Wildl Manage 71:1690–1694
Sarmento P, Cruz J, Eira C, Fonseca C (2009) Spatial colonization by feral domestic cats Felis catus of former wildcat Felis silvestris silvestris home ranges. Acta Theriol 54:31–38
Taberlet P, Waits LP, Luikart G (1999) Noninvasive genetic sampling: look before you leap. Trends Ecol Evol 14:323–327
Teerink BJ (1991) Atlas and identification key hair of West-European mammals. Cambridge University Press, Cambridge
Thompson JD, Higgins DG, Gibson TJ (1994) Clustal-W—improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res 22:4673–4680
Toth AM (2002) Identification of Hungarian Mustelidae and other small carnivores using guard hair analysis. Acta Zool Acad Sci Hung 48:237–250
Valière N (2002) GIMLET: a computer program for analysing genetic individual identification data. Mol Ecol Notes 10:1046–1046
Waits LP, Paetkau D (2005) Noninvasive genetic sampling tools for wildlife biologists: a review of applications and recommendations for accurate data collection. J Wildl Manage 69:1419–1433
Acknowledgements
We thank Achim Frede and the team from the Nationalpark Kellerwald-Edersee and Karsten Wittern (Förderverein für den Nationalpark Kellerwald-Edersee e.V.). We are also grateful to numerous wildcat experts in Germany and Switzerland who helped to develop the methodology shown in this manuscript, including Thomas Mölich, Burkhard Vogel and Thomas Norgall (Bund für Umwelt- und Naturschutz Deutschland, BUND); Jürgen Thein, Karsten Hupe, Malte Götz, Martina Denk and Tabea Stoeckle. Parts of the applied molecular methods were originally developed by ECOGENICS, CH. Wildcat tissue samples were kindly provided by Mathias Herrmann, Matthias Krüger, Franz Müller, Uwe Müller and Jacques Pir. Technical assistance in the laboratory by Mascha Siemund and João Barateiro Diogo is gratefully acknowledged. We appreciate the work of Armin Bürgel, who was an encouraged supporter of this project. This study was funded by the Kellerwald-Edersee National Park. Additional funding comes from the BUND Hessen and the Landesoffensive zur Entwicklung wissenschaftlich-ökonomischer Exzellenz of the state of Hesse. RHSK was funded by grant SAW-2011-SGN-3 from the Leibniz Association.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by P. C. Alves
Rights and permissions
About this article
Cite this article
Steyer, K., Simon, O., Kraus, R.H.S. et al. Hair trapping with valerian-treated lure sticks as a tool for genetic wildcat monitoring in low-density habitats. Eur J Wildl Res 59, 39–46 (2013). https://doi.org/10.1007/s10344-012-0644-0
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10344-012-0644-0