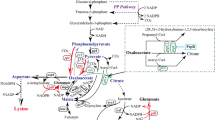
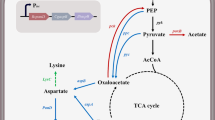
Abstract
Traditional amino acid producers typically exhibit the low glucose uptake rate and growth deficiency, resulting in a long fermentation time because of the accumulation of side mutations in breeding of strains. In this study, we demonstrate that the efficiency of l-lysine production in traditional l-lysine producer Corynebacterium glutamicum ZL-9 can be improved by rationally engineering glucose uptake systems. To do this, different bypasses for glucose uptake were investigated to reveal the best glucose uptake system for l-lysine production in traditional l-lysine producer. This study showed that overexpression of the key genes in PTSGlc or non-PTSGlc increased the glucose consumption, growth rate, and l-lysine production. However, increasing the function of PTSGlc in glucose uptake led to the increase of by-products, especially for plasmid-mediated expression system. Increasing the participation of non-PTSGlc in glucose utilization showed the best glucose uptake system for l-lysine production. The final strain ZL-92 with increasing the expression level of iolT1, iolT2 and ppgK could produce 201.6 ± 13.8 g/L of l-lysine with a productivity of 5.04 g/L/h and carbon yield of 0.65 g/(g glucose) in fed-batch culture. This is the first report of a rational modification of glucose uptake systems that improve the efficiency of l-lysine production through increasing the participation of non-PTSGlc in glucose utilization in traditional l-lysine producer. Similar strategies can be also used for producing other amino acids or their derivatives.





Similar content being viewed by others
References
Xu JZ, Zhang JL, Guo YF, Zai YG, Zhang WG (2013) Improvement of cell growth and l-lysine production by genetically modified Corynebacterium glutamicum during growth on molasses. J Ind Microbiol Biotechnol 40:1423–1432. https://doi.org/10.1007/s10295-013-1329-8
Sagong HY, Kim KJ (2017) Structural basis for redox sensitivity in Corynebacterium glutamicum diaminopimelate epimerase: an enzyme involved in l-lysine biosynthesis. Sci Rep UK 7:42318–42330. https://doi.org/10.1038/srep42318
Xu JZ, Yang HK, Liu LM, Wang YY, Zhang WG (2018) Rational modification of Corynebacterium glutamicum dihydrodipicolinate reductase to switch the nucleotide-cofactor specificity for increasing l-lysine production. Biotechnol Bioeng 115:1764–1777. https://doi.org/10.1002/bit.26591
Irshad S, Faisal M, Hashmi AS, Javed MM, Baber ME, Awan AR, Anjum AA (2015) Mass production and recovery of l-lysine by microbial fermentation using Brevibacterium flavum. J Anim Plant Sci 25:290–294
Xu JZ, Wu ZH, Gao SJ, Zhang WG (2018) Rational modification of tricarboxylic acid cycle for improving l-lysine production in Corynebacterium glutamicum. Microb Cell Fact 17:105–118. https://doi.org/10.1186/s12934-018-0958-z
Sgobba E, Stumpf AK, Vortmann M, Jagmann N, Krehenbrink M, Dirks-Hofmeister ME, Moerschbacher B, Philipp B, Wendisch VF (2018) Synthetic Escherichia coli-Corynebacterium glutamicum consortia for l-lysine production from starch and sucrose. Biores Technol 260:302–310. https://doi.org/10.1016/j.biortech.2018.03.113
Wang Y, Li QG, Zheng P, Guo YM, Wang LX, Zhang TC, Sun JB, Ma YH (2016) Evolving the l-lysine high-producing strain of Escherichia coli using a newly developed high-throughput screening method. J Ind Microbiol Biotechnol 43:1227–1235. https://doi.org/10.1007/s10295-016-1803-1
Ishikawa K, Toda-Murakoshi Y, Ohnishi F, Kondo K, Osumi T, Asano K (2008) Medium composition suitable for l-lysine production by Methylophilus methylotrophus in fed-batch cultivation. J Biosci Bioeng 106:574–579. https://doi.org/10.1263/jbb.106.574
Akyilmaz E, Erdogan A, Ozturk R, Yasa I (2007) Sensitive determination of l-lysine with a new amperometric microbial biosensor based on Saccharomyces cerevisiae yeast cells. Biosens Bioelectron 22:1055–1060. https://doi.org/10.1016/j.bios.2006.04.023
Bhattacharyya R, Samanta TK (1992) l-lysine production by double auxotrophic and AEC resistant mutants of Arthrobacter globiformis Tr9. Res Ind 37:1–6
Wittmann C, Becker J (2007) The l-lysine story: from metabolic pathways to industrial production. In: Wendisch VF (ed) Amino Acid Biosynthesis. Springer, Berlin Heidelberg
Xu JZ, Zhang JL, Liu DD, Zhang WG (2016) Increased glucose utilization and cell growth of Corynebacterium glutamicum by modifying the glucose-specific phosphotransferase system (PTSGlc) genes. Can J Microbiol 62:983–992. https://doi.org/10.1139/cjm-2016-0027
Becker J, Wittmann C (2012) Bio-based production of chemicals, materials and fuels - Corynebacterium glutamicum as versatile cell factory. Curr Opin Biotechnol 23:631–640. https://doi.org/10.1016/j.copbio.2011.11.012
Hasegawa S, Tanaka Y, Suda M, Jojima T, Inui M (2017) Enhanced glucose consumption and organic acid production by engineered Corynebacterium glutamicum based on analysis of a pfkB1 deletion mutant. Appl Environ Microbiol. https://doi.org/10.1128/aem.02638-16
Xu JZ, Yang HK, Zhang WG (2018) NADPH metabolism: a survey of its theoretical characteristics and manipulation strategies in amino acid biosynthesis. Crit Rev Biotechnol 38:1061–1076. https://doi.org/10.1080/07388551.2018.1437387
Ikeda M, Noguchi N, Ohshita M, Senoo A, Mitsuhashi S, Takeno S (2015) A third glucose uptake bypass in Corynebacterium glutamicum ATCC 31833. Appl Microbiol Biotechnol 99:2741–2750. https://doi.org/10.1007/s00253-014-6323-1
Ikeda M, Mizuno Y, Awane S, Hayashi M, Mitsuhashi S, Takeno S (2011) Identification and application of a different glucose uptake system that functions as an alternative to the phosphotransferase system in Corynebacterium glutamicum. Appl Microbiol Biotechnol 90:1443–1451. https://doi.org/10.1007/s00253-011-3210-x
Lindner SN, Seibold GM, Kramer R, Wendisch VF (2011) Impact of a new glucose utilization pathway in amino acid-producing Corynebacterium glutamicum. Bioeng Bugs 2:291–295. https://doi.org/10.4161/bbug.2.5.17116
CocaignBousquet M, Guyonvarch A, Lindley ND (1996) Growth rate-dependent modulation of carbon flux through central metabolism and the kinetic consequences for glucose-limited chemostat cultures of Corynebacterium glutamicum. Appl Environ Microbiol 62:429–436
Tanaka Y, Okai N, Teramoto H, Inui M, Yukawa H (2008) Regulation of the expression of phosphoenolpyruvate: carbohydrate phosphotransferase system (PTS) genes in Corynebacterium glutamicum R. Microbiol SGM 154:264–274. https://doi.org/10.1099/mic.0.2007/008862-0
Petersen S, Mack C, de Graaf AA, Riedel C, Eikmanns BJ, Sahm H (2001) Metabolic consequences of altered phosphoenolpyruvate carboxykinase activity in Corynebacterium glutamicum reveal anaplerotic regulation mechanisms in vivo. Metab Eng 3:344–361. https://doi.org/10.1006/mben.2001.0198
Lindner SN, Seibold GM, Henrich A, Kramer R, Wendisch VF (2011) Phosphotransferase system-independent glucose utilization in Corynebacterium glutamicum by inositol permeases and glucokinases. Appl Environ Microbiol 77:3571–3581. https://doi.org/10.1128/AEM.02713-10
Ikeda M (2012) Sugar transport systems in Corynebacterium glutamicum: features and applications to strain development. Appl Microbiol Biotechnol 96:1191–1200. https://doi.org/10.1007/s00253-012-4488-z
Becker J, Zelder O, Hafner S, Schroder H, Wittmann C (2011) From zero to hero–design-based systems metabolic engineering of Corynebacterium glutamicum for l-lysine production. Metab Eng 13:159–168. https://doi.org/10.1016/j.ymben.2011.01.003
Blombach B, Arndt A, Auchter M, Eikmanns BJ (2009) L-valine production during growth of pyruvate dehydrogenase complex-deficient Corynebacterium glutamicum in the presence of ethanol or by inactivation of the transcriptional regulator SugR. Appl Environ Microbiol 75:1197–1200. https://doi.org/10.1128/AEM.02351-08
Dominguez H, CocaignBousquet M, Lindley ND (1997) Simultaneous consumption of glucose and fructose from sugar mixtures during botch growth of Corynebacterium glutamicum. Appl Microbiol and Biotechnol 47:600–603. https://doi.org/10.1007/s002530050980
van der Rest ME, Lange C, Molenaar D (1999) A heat shock following electroporation induces highly efficient transformation of Corynebacterium glutamicum with xenogeneic plasmid DNA. Appl Microbiol Biotechnol 52:541–545. https://doi.org/10.1007/s002530051557
Xu JZ, Han M, Zhang JL, Guo YF, Zhang WG (2014) Metabolic engineering Corynebacterium glutamicum for the l-lysine production by increasing the flux into l-lysine biosynthetic pathway. Amino Acids 46:2165–2175. https://doi.org/10.1007/s00726-014-1768-1
Yamamoto S, Gunji W, Suzuki H, Toda H, Suda M, Jojima T, Inui M, Yukawa H (2012) Overexpression of genes encoding glycolytic enzymes in Corynebacterium glutamicum enhances glucose metabolism and alanine production under oxygen deprivation conditions. Appl Environ Microbiol 78:4447–4457. https://doi.org/10.1128/AEM.07998-11
Xu JZ, Han M, Zhang JL, Guo YF, Qian H, Zhang WG (2014) Improvement of l-lysine production combines with minimization of by-products synthesis in Corynebacterium glutamicum. J Chem Technol Biotechnol 89:1924–1933. https://doi.org/10.1002/jctb.4278
Blombach B, Seibold GM (2010) Carbohydrate metabolism in Corynebacterium glutamicum and applications for the metabolic engineering of l-lysine production strains. Appl Microbiol Biotechnol 86:1313–1322. https://doi.org/10.1007/s00253-010-2537-z
Wang C, Cai H, Zhou ZH, Zhang K, Chen ZJ, Chen YL, Wan HG, Ouyang PK (2014) Investigation of ptsG gene in response to xylose utilization in Corynebacterium glutamicum. J Ind Microbiol Biotechnol 41:1249–1258. https://doi.org/10.1007/s10295-014-1455-y
Clark DP, Pazdernik NJ (2016) Protein Engineering. In: Clark DP, Pazdernik NJ (eds) Biotechnology. Academic Cell, Burlingon
Engels V, Wendisch VF (2007) The DeoR-type regulator SugR represses expression of ptsG in Corynebacterium glutamicum. J Bacteriol 189:2955–2966. https://doi.org/10.1128/JB.01596-06
Krause FS, Henrich A, Blombach B, Kramer R, Eikmanns BJ, Seibold GM (2010) Increased glucose utilization in Corynebacterium glutamicum by use of maltose, and its application for the improvement of l-valine productivity. Appl Environ Microbiol 76:370–374. https://doi.org/10.1128/AEM.01553-09
Lara AR, Caspeta L, Gosset G, Bolivar F, Ramirez OT (2008) Utility of an Escherichia coli strain engineered in the substrate uptake system for improved culture performance at high glucose and cell concentrations: an fed-batch cultures. Biotechnol Bioeng 99:893–901. https://doi.org/10.1002/bit.21664
Ikeda M, Katsumata R (1998) A novel system with positive selection for the chromosomal integration of replicative plasmid DNA in Corynebacterium glutamicum. Microbiology SGM 144:1863–1868. https://doi.org/10.1099/00221287-144-7-1863
Kang CW, Lim HG, Yang J, Noh MH, Seo SW, Jung GY (2018) Synthetic auxotrophs for stable and tunable maintenance of plasmid copy number. Metab Eng 48:121–128. https://doi.org/10.1016/j.ymben.2018.05.020
Klaffl S, Brocker M, Kalinowski J, Eikmanns BJ, Bott M (2013) Complex regulation of the phosphoenolpyruvate carboxykinase gene pck and characterization ofits GntR-type regulator IolR as a repressor of myo-inositol utilization genes in Corynebacterium glutamicum. J Bacteriol 195:4283–4296. https://doi.org/10.1128/JB.00265-13
Brusseler C, Radek A, Tenhaef N, Krumbach K, Noack S, Marienhagen J (2018) The myo-inositol/proton symporter IolT1 contributes to d-xylose uptake in Corynebacterium glutamicum. Biores Technol 249:953–961. https://doi.org/10.1016/j.biortech.2017.10.098
Sawada K, Zen-in S, Wada M, Yokota A (2010) Metabolic changes in a pyruvate kinase gene deletion mutant of Corynebacterium glutamicum ATCC 13032. Metab Eng 12:401–407. https://doi.org/10.1016/j.ymben.2010.01.004
Becker J, Klopprogge C (2008) Wittmann C (2008) Metabolic responses to pyruvate kinase deletion in lysine producing Corynebacterium glutamicum. Microb Cell Fact 7:8. https://doi.org/10.1186/1475-2859-7-8
Schafer A, Tauch A, Jager W, Kalinowski J, Thierbach G, Puhler A (1994) Small mobilizable multipurpose cloning vectors derived from the Escherichia coli plasmids pK18 and pK19—selection of defined deletions in the chromosome of Corynebacteriumglutamicum. Gene 145:69–73
Ohnishi J, Ikeda M (2006) Comparisons of potentials for l-lysine production among different Corynebacterium glutamicum strains. Biosci Biotechnol Biochem 70:1017–1020. https://doi.org/10.1271/bbb.70.1017
van Ooyen J, Noack S, Bott M, Reth A, Eggeling L (2012) Improved l-lysine production with Corynebacterium glutamicum and systemic insight into citrate synthase flux and activity. Biotechnol Bioeng 109:2070–2081. https://doi.org/10.1002/bit.24486
Sindelar G, Wendisch VF (2007) Improving lysine production by Corynebacterium glutamicum through DNA microarray-based identification of novel target genes. Appl Microbiol Biotechnol 76:677–689. https://doi.org/10.1007/s00253-007-0916-x
Acknowledgements
This work was supported by the National Natural Science Foundation of China [Grant Number 31601459], the China Postdoctoral Science Foundation [Grant Number 2016M590410], and the National First-class Discipline Program of Light Industry Technology and Engineering [Grant Number LITE2018-07]. We thank Dr. XL Chen (State Key Laboratory of Food Science and Technology at Jiangnan University) for assistance in data analysis.
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Xu, JZ., Yu, HB., Han, M. et al. Metabolic engineering of glucose uptake systems in Corynebacterium glutamicum for improving the efficiency of l-lysine production. J Ind Microbiol Biotechnol 46, 937–949 (2019). https://doi.org/10.1007/s10295-019-02170-w
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10295-019-02170-w