Abstract
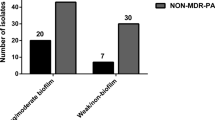
Pseudomonas aeruginosa (Pa) is one of the major bacterial pathogens causing nosocomial infections. During the past few decades, multidrug-resistant (MDR) and extensively drug-resistant (XDR) lineages of Pa have emerged in hospital settings with increasing numbers. However, it remains unclear which determinants of Pa facilitated this spread. A total of 211 clinical XDR and 38 susceptible clinical Pa isolates (nonXDR), as well as 47 environmental isolates (EI), were collected at the Heidelberg University Hospital. We used RAPD PCR to identify genetic clusters. Carriage of carbapenamases (CPM) and virulence genes were analyzed by PCR, biofilm formation capacity was assessed, in vitro fitness was evaluated using competitive growth assays, and interaction with the host’s immune system was analyzed using serum killing and neutrophil killing assays. XDR isolates showed significantly elevated biofilm formation (p < 0.05) and higher competitive fitness compared to nonXDR and EI isolates. Thirty percent (62/205) of the XDR isolates carried a CPM. Similarities in distribution of virulence factors, as well as biofilm formation properties, between CPM+ Pa isolates and EI and between CPM- and nonXDR isolates were detected. Molecular typing revealed two distinct genetic clusters within the XDR population, which were characterized by even higher biofilm formation. In contrast, XDR isolates were more susceptible to the immune response than nonXDR isolates. Our study provides evidence that the ability to form biofilms is an outstanding determinant for persistence and endemic spread of Pa in the hospital setting.





Similar content being viewed by others
References
Livermore DM (2002) Multiple mechanisms of antimicrobial resistance in Pseudomonas aeruginosa: our worst nightmare? Clin Infect Dis Off Publ Infect Dis Soc Am 34(5):634–640. doi:10.1086/338782
Oliver A, Mulet X, Lopez-Causape C, Juan C (2015) The increasing threat of Pseudomonas aeruginosa high-risk clones. Drug Resistance Updat Rev Comment Antimicrob Anticancer Chemother 21–22:41–59. doi:10.1016/j.drup.2015.08.002
Potron A, Poirel L, Nordmann P (2015) Emerging broad-spectrum resistance in Pseudomonas aeruginosa and Acinetobacter baumannii: mechanisms and epidemiology. Int J Antimicrob Agents 45(6):568–585. doi:10.1016/j.ijantimicag.2015.03.001
Morita Y, Tomida J, Kawamura Y (2014) Responses of Pseudomonas aeruginosa to antimicrobials. Front Microbiol 4:422. doi:10.3389/fmicb.2013.00422
Hilker R, Munder A, Klockgether J, Losada PM, Chouvarine P, Cramer N, Davenport CF, Dethlefsen S, Fischer S, Peng H, Schonfelder T, Turk O, Wiehlmann L, Wolbeling F, Gulbins E, Goesmann A, Tummler B (2015) Interclonal gradient of virulence in the Pseudomonas aeruginosa pangenome from disease and environment. Environ Microbiol 17(1):29–46. doi:10.1111/1462-2920.12606
Dubern JF, Cigana C, De Simone M, Lazenby J, Juhas M, Schwager S, Bianconi I, Doring G, Eberl L, Williams P, Bragonzi A, Camara M (2015) Integrated whole-genome screening for Pseudomonas aeruginosa virulence genes using multiple disease models reveals that pathogenicity is host specific. Environ Microbiol 17(11):4379–4393. doi:10.1111/1462-2920.12863
Stoodley P, Sauer K, Davies DG, Costerton JW (2002) Biofilms as complex differentiated communities. Annu Rev Microbiol 56:187–209. doi:10.1146/annurev.micro.56.012302.160705
Laverty G, Gorman SP, Gilmore BF (2014) Biomolecular mechanisms of Pseudomonas aeruginosa and escherichia coli biofilm formation. Pathogens 3(3):596–632. doi:10.3390/pathogens3030596
Wang S, Yu S, Zhang Z, Wei Q, Yan L, Ai G, Liu H, Ma LZ (2014) Coordination of swarming motility, biosurfactant synthesis, and biofilm matrix exopolysaccharide production in Pseudomonas aeruginosa. Appl Environ Microbiol 80(21):6724–6732. doi:10.1128/AEM.01237-14
Franklin MJ, Nivens DE, Weadge JT, Howell PL (2011) Biosynthesis of the Pseudomonas aeruginosa extracellular polysaccharides, Alginate, Pel, and Psl. Front Microbiol 2:167. doi:10.3389/fmicb.2011.00167
Ma L, Wang J, Wang S, Anderson EM, Lam JS, Parsek MR, Wozniak DJ (2012) Synthesis of multiple Pseudomonas aeruginosa biofilm matrix exopolysaccharides is post-transcriptionally regulated. Environ Microbiol 14(8):1995–2005. doi:10.1111/j.1462-2920.2012.02753.x
Oliveira NM, Martinez-Garcia E, Xavier J, Durham WM, Kolter R, Kim W, Foster KR (2015) Biofilm formation as a response to ecological competition. PLoS Biol 13(7), e1002191. doi:10.1371/journal.pbio.1002191
Walker TS, Tomlin KL, Worthen GS, Poch KR, Lieber JG, Saavedra MT, Fessler MB, Malcolm KC, Vasil ML, Nick JA (2005) Enhanced Pseudomonas aeruginosa biofilm development mediated by human neutrophils. Infect Immun 73(6):3693–3701. doi:10.1128/IAI.73.6.3693-3701.2005
Costerton JW, Stewart PS, Greenberg EP (1999) Bacterial biofilms: a common cause of persistent infections. Science 284(5418):1318–1322
Hansch GM, Brenner-Weiss G, Prior B, Wagner C, Obst U (2008) The extracellular polymer substance of Pseudomonas aeruginosa: too slippery for neutrophils to migrate on? Int J Artif Organs 31(9):796–803
Gunther F, Wabnitz GH, Stroh P, Prior B, Obst U, Samstag Y, Wagner C, Hansch GM (2009) Host defence against Staphylococcus aureus biofilms infection: phagocytosis of biofilms by polymorphonuclear neutrophils (PMN). Mol Immunol 46(8–9):1805–1813. doi:10.1016/j.molimm.2009.01.020
Meyle E, Stroh P, Gunther F, Hoppy-Tichy T, Wagner C, Hansch GM (2010) Destruction of bacterial biofilms by polymorphonuclear neutrophils: relative contribution of phagocytosis, DNA release, and degranulation. Int J Artif Organs 33(9):608–620
Savoia D, Deplano C, Zucca M (2008) Pseudomonas aeruginosa and Burkholderia cenocepacia infections in patients affected by cystic fibrosis: serum resistance and antibody response. Immunol Investig 37(1):19–27. doi:10.1080/08820130701741775
Mueller-Ortiz SL, Drouin SM, Wetsel RA (2004) The alternative activation pathway and complement component C3 are critical for a protective immune response against Pseudomonas aeruginosa in a murine model of pneumonia. Infect Immun 72(5):2899–2906
Berends ET, Kuipers A, Ravesloot MM, Urbanus RT, Rooijakkers SH (2014) Bacteria under stress by complement and coagulation. FEMS Microbiol Rev 38(6):1146–1171. doi:10.1111/1574-6976.12080
Klockgether J, Cramer N, Wiehlmann L, Davenport CF, Tummler B (2011) Pseudomonas aeruginosa genomic structure and diversity. Front Microbiol 2:150. doi:10.3389/fmicb.2011.00150
Lee DG, Urbach JM, Wu G, Liberati NT, Feinbaum RL, Miyata S, Diggins LT, He J, Saucier M, Deziel E, Friedman L, Li L, Grills G, Montgomery K, Kucherlapati R, Rahme LG, Ausubel FM (2006) Genomic analysis reveals that Pseudomonas aeruginosa virulence is combinatorial. Genome Biol 7(10):R90. doi:10.1186/gb-2006-7-10-r90
Soong G, Muir A, Gomez MI, Waks J, Reddy B, Planet P, Singh PK, Kaneko Y, Wolfgang MC, Hsiao YS, Tong L, Prince A (2006) Bacterial neuraminidase facilitates mucosal infection by participating in biofilm production. J Clin Invest 116(8):2297–2305. doi:10.1172/JCI27920
Michalska M, Wolf P (2015) Pseudomonas Exotoxin A: optimized by evolution for effective killing. Front Microbiol 6:963. doi:10.3389/fmicb.2015.00963
Ballok AE, O’Toole GA (2013) Pouring salt on a wound: Pseudomonas aeruginosa virulence factors alter Na + and Cl- flux in the lung. J Bacteriol 195(18):4013–4019. doi:10.1128/JB.00339-13
Sawa T (2014) The molecular mechanism of acute lung injury caused by Pseudomonas aeruginosa: from bacterial pathogenesis to host response. J Intensive Care 2(1):10. doi:10.1186/2052-0492-2-10
Allewelt M, Coleman FT, Grout M, Priebe GP, Pier GB (2000) Acquisition of expression of the Pseudomonas aeruginosa ExoU cytotoxin leads to increased bacterial virulence in a murine model of acute pneumonia and systemic spread. Infect Immun 68(7):3998–4004
Yahr TL, Hovey AK, Kulich SM, Frank DW (1995) Transcriptional analysis of the Pseudomonas aeruginosa exoenzyme S structural gene. J Bacteriol 177(5):1169–1178
Vance RE, Rietsch A, Mekalanos JJ (2005) Role of the type III secreted exoenzymes S, T, and Y in systemic spread of Pseudomonas aeruginosa PAO1 in vivo. Infect Immun 73(3):1706–1713. doi:10.1128/IAI.73.3.1706-1713.2005
Stevens TC, Ochoa CD, Morrow KA, Robson MJ, Prasain N, Zhou C, Alvarez DF, Frank DW, Balczon R, Stevens T (2014) The Pseudomonas aeruginosa exoenzyme Y impairs endothelial cell proliferation and vascular repair following lung injury. Am J Physiol Lung Cell Mol Physiol 306(10):L915–L924. doi:10.1152/ajplung.00135.2013
Morrow KA, Ochoa CD, Balczon R, Zhou C, Cauthen L, Alexeyev M, Schmalzer KM, Frank DW, Stevens T (2016) Pseudomonas aeruginosa exoenzymes U and Y induce a transmissible endothelial proteinopathy. Am J Physiol Lung Cell Mol Physiol 310(4):L337–L353. doi:10.1152/ajplung.00103.2015
Magiorakos AP, Srinivasan A, Carey RB, Carmeli Y, Falagas ME, Giske CG, Harbarth S, Hindler JF, Kahlmeter G, Olsson-Liljequist B, Paterson DL, Rice LB, Stelling J, Struelens MJ, Vatopoulos A, Weber JT, Monnet DL (2012) Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: an international expert proposal for interim standard definitions for acquired resistance. Clin Microbiol Infect 18(3):268–281. doi:10.1111/j.1469-0691.2011.03570.x
He J, Baldini RL, Deziel E, Saucier M, Zhang Q, Liberati NT, Lee D, Urbach J, Goodman HM, Rahme LG (2004) The broad host range pathogen Pseudomonas aeruginosa strain PA14 carries two pathogenicity islands harboring plant and animal virulence genes. Proc Natl Acad Sci USA 101(8):2530–2535
Kukavica-Ibrulj I, Bragonzi A, Paroni M, Winstanley C, Sanschagrin F, O’Toole GA, Levesque RC (2008) In vivo growth of Pseudomonas aeruginosa strains PAO1 and PA14 and the hypervirulent strain LESB58 in a rat model of chronic lung infection. J Bacteriol 190(8):2804–2813. doi:10.1128/JB.01572-07
Campbell M, Mahenthiralingam E, Speert DP (2000) Evaluation of random amplified polymorphic DNA typing of Pseudomonas aeruginosa. J Clin Microbiol 38(12):4614–4615
O’Toole GA (2011) Microtiter dish biofilm formation assay. J Vis Exp JoVE (47). doi:10.3791/2437
Kaszab E, Szoboszlay S, Dobolyi C, Hahn J, Pek N, Kriszt B (2011) Antibiotic resistance profiles and virulence markers of Pseudomonas aeruginosa strains isolated from composts. Bioresour Technol 102(2):1543–1548. doi:10.1016/j.biortech.2010.08.027
Sun Z, Jiao X, Peng Q, Jiang F, Huang Y, Zhang J, Yao F (2013) Antibiotic resistance in Pseudomonas aeruginosa is associated with decreased fitness. Cell Physiol Biochem Int J Exp Cell Physiol Biochem Pharmacol 31(2–3):347–354. doi:10.1159/000343372
Sanchez P, Linares JF, Ruiz-Diez B, Campanario E, Navas A, Baquero F, Martinez JL (2002) Fitness of in vitro selected Pseudomonas aeruginosa nalB and nfxB multidrug resistant mutants. J Antimicrob Chemother 50(5):657–664
Olivares J, Alvarez-Ortega C, Martinez JL (2014) Metabolic compensation of fitness costs associated with overexpression of the multidrug efflux pump MexEF-OprN in Pseudomonas aeruginosa. Antimicrob Agents Chemother 58(7):3904–3913. doi:10.1128/AAC.00121-14
Kugelberg E, Lofmark S, Wretlind B, Andersson DI (2005) Reduction of the fitness burden of quinolone resistance in Pseudomonas aeruginosa. J Antimicrob Chemother 55(1):22–30. doi:10.1093/jac/dkh505
Donlan RM, Costerton JW (2002) Biofilms: survival mechanisms of clinically relevant microorganisms. Clin Microbiol Rev 15(2):167–193
Mulet X, Cabot G, Ocampo-Sosa AA, Dominguez MA, Zamorano L, Juan C, Tubau F, Rodriguez C, Moya B, Pena C, Martinez-Martinez L, Oliver A, Spanish Network for Research in Infectious D (2013) Biological markers of Pseudomonas aeruginosa epidemic high-risk clones. Antimicrob Agents Chemother 57(11):5527–5535. doi:10.1128/AAC.01481-13
Taccone FS, Cotton F, Roisin S, Vincent JL, Jacobs F (2012) Optimal meropenem concentrations to treat multidrug-resistant Pseudomonas aeruginosa septic shock. Antimicrob Agents Chemother 56(4):2129–2131. doi:10.1128/AAC.06389-11
Ghazi IM, Crandon JL, Lesho EP, McGann P, Nicolau DP (2015) Efficacy of humanized high-dose meropenem, cefepime, and levofloxacin against Enterobacteriaceae isolates producing Verona integron-encoded metallo-beta-lactamase (VIM) in a murine thigh infection model. Antimicrob Agents Chemother 59(11):7145–7147. doi:10.1128/AAC.00794-15
Martinez-Ramos I, Mulet X, Moya B, Barbier M, Oliver A, Alberti S (2014) Overexpression of MexCD-OprJ reduces Pseudomonas aeruginosa virulence by increasing its susceptibility to complement-mediated killing. Antimicrob Agents Chemother 58(4):2426–2429. doi:10.1128/AAC.02012-13
Schiller NL, Joiner KA (1986) Interaction of complement with serum-sensitive and serum-resistant strains of Pseudomonas aeruginosa. Infect Immun 54(3):689–694
She P, Chen L, Qi Y, Xu H, Liu Y, Wang Y, Luo Z, Wu Y (2016) Effects of human serum and apo-Transferrin on Staphylococcus epidermidis RP62A biofilm formation. Microbiologyopen. doi:10.1002/mbo3.379
Cho HH, Kwon KC, Kim S, Koo SH (2014) Correlation between virulence genotype and fluoroquinolone resistance in carbapenem-resistant Pseudomonas aeruginosa. Ann Lab Med 34(4):286–292. doi:10.3343/alm.2014.34.4.286
Kulasekara BR, Kulasekara HD, Wolfgang MC, Stevens L, Frank DW, Lory S (2006) Acquisition and evolution of the exoU locus in Pseudomonas aeruginosa. J Bacteriol 188(11):4037–4050. doi:10.1128/JB.02000-05
Pena C, Cabot G, Gomez-Zorrilla S, Zamorano L, Ocampo-Sosa A, Murillas J, Almirante B, Pomar V, Aguilar M, Granados A, Calbo E, Rodriguez-Bano J, Rodriguez-Lopez F, Tubau F, Martinez-Martinez L, Oliver A, Spanish Network for Research in Infectious D (2015) Influence of virulence genotype and resistance profile in the mortality of Pseudomonas aeruginosa bloodstream infections. Clin Infect Dis Off Publ Inf Dis Soc Am 60(4):539–548. doi:10.1093/cid/ciu866
Sawa T, Shimizu M, Moriyama K, Wiener-Kronish JP (2014) Association between Pseudomonas aeruginosa type III secretion, antibiotic resistance, and clinical outcome: a review. Crit Care 18(6):668. doi:10.1186/s13054-014-0668-9
Wareham DW, Curtis MA (2007) A genotypic and phenotypic comparison of type III secretion profiles of Pseudomonas aeruginosa cystic fibrosis and bacteremia isolates. Int J Med Microbiol IJMM 297(4):227–234. doi:10.1016/j.ijmm.2007.02.004
Feltman H, Schulert G, Khan S, Jain M, Peterson L, Hauser AR (2001) Prevalence of type III secretion genes in clinical and environmental isolates of Pseudomonas aeruginosa. Microbiology 147(Pt 10):2659–2669. doi:10.1099/00221287-147-10-2659
Bradbury RS, Reid DW, Inglis TJ, Champion AC (2011) Decreased virulence of cystic fibrosis Pseudomonas aeruginosa in Dictyostelium discoideum. Microbiol Immunol 55(4):224–230. doi:10.1111/j.1348-0421.2011.00314.x
Hogardt M, Heesemann J (2010) Adaptation of Pseudomonas aeruginosa during persistence in the cystic fibrosis lung. Int J Med Microbiol IJMM 300(8):557–562. doi:10.1016/j.ijmm.2010.08.008
Vidya P, Smith L, Beaudoin T, Yau YC, Clark S, Coburn B, Guttman DS, Hwang DM, Waters V (2016) Chronic infection phenotypes of Pseudomonas aeruginosa are associated with failure of eradication in children with cystic fibrosis. Eur J Clin Microbiol Infect Dis 35(1):67–74. doi:10.1007/s10096-015-2509-4
Rau MH, Hansen SK, Johansen HK, Thomsen LE, Workman CT, Nielsen KF, Jelsbak L, Hoiby N, Yang L, Molin S (2010) Early adaptive developments of Pseudomonas aeruginosa after the transition from life in the environment to persistent colonization in the airways of human cystic fibrosis hosts. Environ Microbiol 12(6):1643–1658. doi:10.1111/j.1462-2920.2010.02211.x
Jones AM, Govan JR, Doherty CJ, Dodd ME, Isalska BJ, Stanbridge TN, Webb AK (2003) Identification of airborne dissemination of epidemic multiresistant strains of Pseudomonas aeruginosa at a CF centre during a cross infection outbreak. Thorax 58(6):525–527
Wainwright CE, France MW, O’Rourke P, Anuj S, Kidd TJ, Nissen MD, Sloots TP, Coulter C, Ristovski Z, Hargreaves M, Rose BR, Harbour C, Bell SC, Fennelly KP (2009) Cough-generated aerosols of Pseudomonas aeruginosa and other Gram-negative bacteria from patients with cystic fibrosis. Thorax 64(11):926–931. doi:10.1136/thx.2008.112466
Willmann M, Bezdan D, Zapata L, Susak H, Vogel W, Schroppel K, Liese J, Weidenmaier C, Autenrieth IB, Ossowski S, Peter S (2015) Analysis of a long-term outbreak of XDR Pseudomonas aeruginosa: a molecular epidemiological study. J Antimicrob Chemother 70(5):1322–1330. doi:10.1093/jac/dku546
Inglis TJ, Benson KA, O’Reilly L, Bradbury R, Hodge M, Speers D, Heath CH (2010) Emergence of multi-resistant Pseudomonas aeruginosa in a Western Australian hospital. J Hosp Infect 76(1):60–65. doi:10.1016/j.jhin.2010.01.026
Wolfgang MC, Kulasekara BR, Liang X, Boyd D, Wu K, Yang Q, Miyada CG, Lory S (2003) Conservation of genome content and virulence determinants among clinical and environmental isolates of Pseudomonas aeruginosa. Proc Natl Acad Sci USA 100(14):8484–8489. doi:10.1073/pnas.0832438100
Shi H, Trinh Q, Xu W, Zhai B, Luo Y, Huang K (2012) A universal primer multiplex PCR method for typing of toxinogenic Pseudomonas aeruginosa. Appl Microbiol Biotechnol 95(6):1579–1587. doi:10.1007/s00253-012-4277-8
Bradbury RS, Roddam LF, Merritt A, Reid DW, Champion AC (2010) Virulence gene distribution in clinical, nosocomial and environmental isolates of Pseudomonas aeruginosa. J Med Microbiol 59(Pt 8):881–890. doi:10.1099/jmm.0.018283-0
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
All authors report no financial or non-financial conflicts of interest. All used isolates were routinely collected in the microbiology laboratory of the Heidelberg University Hospital and stored at −70 °C. The present study thus is descriptive of a bacterial collection of those isolates and additional environmental isolates. Data collected from patients was anonymized and restricted to possible clinical symptoms of infection. Ethical approval and informed consent statements were therefore not required. The study was conducted without additional funding.
Additional information
S. J. Kaiser and N. T. Mutters contributed equally to this work.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Supplement 1
Heat map of the analyzed features for all isolates and groups used in this study. Yellow indicates values close to the median, while red indicates values associated with elevated feature characteristics and green low feature characteristics. (PDF 110 kb)
Rights and permissions
About this article
Cite this article
Kaiser, S.J., Mutters, N.T., DeRosa, A. et al. Determinants for persistence of Pseudomonas aeruginosa in hospitals: interplay between resistance, virulence and biofilm formation. Eur J Clin Microbiol Infect Dis 36, 243–253 (2017). https://doi.org/10.1007/s10096-016-2792-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10096-016-2792-8