Abstract
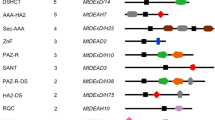
DEAD-box RNA helicase family proteins have been identified in almost all living organisms. Some of them play a crucial role in adaptation to environmental changes and stress response, especially in the low-temperature acclimation in different kinds of organisms. Compared with the full swing study in plants and bacteria, the characters and functions of DEAD-box family proteins had not been surveyed in algae. To identify genes critical for freezing acclimation in algae, we screened DEAD-box RNA helicase genes from the transcriptome sequences of a psychrophilic microalga Chlamydomonas sp. ICE-L which was isolated from Antarctic sea ice. Totally 39 DEAD-box RNA helicase genes had been identified. Most of the DEAD-box RNA helicase have 1:1 homologous relationships in Chlamydomonas reinhardtii and Chlamydomonas sp. ICE-L with several exceptions. The homologous proteins in ICE-L to the helicases critical for cold or freezing tolerance in Arabidopsis thaliana had been identified based on phylogenetic comparison studies. The response of these helicase genes is not always identical in the Chlamydomonas sp. ICE-L and Arabidopsis under the same low-temperature treatment. The expression of several DEAD-box RNA helicase genes including CiRH5, CiRH25, CiRH28, and CiRH55 were significantly up-regulated under freezing treatment of ICE-L and their function in freezing acclimation of ICE-L deserved further investigation.





Similar content being viewed by others
References
Chen ZZ, Xue CH, Zhu S, Zhou F, Xuefeng BL, Liu G, Chen L (2005) GoPipe: streamlined gene ontology annotation for batch anonymous sequences with statistics. Prog Biochem Biophys 32:187–191
Chung E, Cho CW, Yun BH, Choi HK, So HA, Lee SW, Lee JH (2009) Molecular cloning and characterization of the soybean DEAD-box RNA helicase gene induced by low temperature and high salinity stress. Gene 443(1):91–99
Fairman-Williams ME, Guenther UP, Jankowsky E (2010) SF1 and SF2 helicases: family matters. Curr Opin Struct Biol 20:313–324
Gong Z, Lee H, Xiong L, Jagendorf A, Stevenson B, Zhu JK (2002) RNA helicase-like protein as an early regulator of transcription factors for plant chilling and freezing tolerance. Proc Natl Acad Sci USA 99:11507–11512
Gong Z, Dong CH, Lee H et al (2005) A DEAD box RNA helicase is essential for mRNA export and important for development and stress responses in Arabidopsis. Plant Cell 17(1):256–267
Gu L, Xu T, Lee K, Lee KH, Kang H (2014) A chloroplast-localized DEAD-box RNA helicase AtRH3 is essential for intron splicing and plays an important role in the growth and stress response in Arabidopsis thaliana. Plant Physiol Biochem 82:309–318
Guan QM, Wu JM, Zhang YY, Jiang C, Liu R, Chai C, Zhu J (2013) A DEAD box RNA helicase is critical for pre-mRNA splicing, cold-responsive gene regulation, and cold tolerance in Arabidopsis. Plant Cell 25:342–356
Iost I, Bizebard T, Dreyfus M (2013) Functions of DEAD-box proteins in bacteria: current knowledge and pending questions. BBA 1829(8):866–877
Jagessar KL, Jain C (2010) Functional and molecular analysis of Escherichia coli strains lacking multiple DEAD-box helicases. RNA 16:1386–1392
Jankowsky A, Guenther UP, Jankowsky E (2011) The RNA helicase database. Nucleic Acids Res 39:338–341
Kanehisa M, Goto S, Furumichi M, Tanabe M, Hirakawa M (2010) KEGG for representation and analysis of molecular networks involving diseases and drugs. Nucleic Acids Res 38:355–360
Kant P, Kant S, Gordon M, Shaked R, Barak S (2007) STRESS RESPONSE SUPPRESSOR1 and STRESS RESPONSE SUPPRESSOR2, two DEAD-box RNA helicases that attenuate Arabidopsis responses to multiple abiotic stresses. Plant Physiol 145(3):814–830
Kim JS, Kim KA, Oh TR, Park CM, Kang H (2008) Functional characterization of DEAD-box RNA helicases in Arabidopsis thaliana under abiotic stress conditions. Plant Cell Physiol 49(10):1563–1571
Kumar S, Nei M, Dudley J, Tamura K (2008) MEGA: a biologistcentric software for evolutionary analysis of DNA and protein sequences. Brief Bioinform 9(4):299–306
Lehnik-Habrink M, Rempeters L, Kovács ÁT, Wrede C, Baierlein C, Krebber H et al (2013) DEAD-box RNA helicases in Bacillus subtilis have multiple functions and act independently from each other. J Bacteriol 195:534–544
Linder P, Fuller-Pace FV (2013) Looking back on the birth of DEAD-box RNA helicases. BBA 1829:750–755
Liu C, Huang X, Wang X, Zhang X, Li G (2006) Phylogenetic studies on two strains of Antarctic ice algae based on morphological and molecular characteristics. Phycologia 45:190–198
Liu C, Wu G, Huang X, Liu S, Cong B (2012) Validation of housekeeping genes for gene expression studies in an ice alga Chlamydomonas during freezing acclimation. Extremophiles 16:419–425
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C[T]) method. Methods 25:402–408
Markkula A, Mattila M, Lindström M, Korkeala H (2012) Genes encoding putative DEAD-box RNA helicases in Listeria monocytogenes EGD-e are needed for growth and motility at 3 °C. Environ Microbiol 14:2223–2232
Merchant SS, Prochnik SE, Vallon O, Harris EH, Karpowicz SJ, Witman GB et al (2007) The Chlamydomonas genome reveals the evolution of key animal and plant functions. Science 318:245–250
Mingam A, Toffano-Nioche C, Brunaud V, Boudet N, Kreis M, Lecharny A (2004) DEAD-box RNA helicases in Arabidopsis thaliana: establishing a link between quantitative expression, gene structure and evolution of a family of genes. Plant Biotechnol J 2:401–415
Mock T, Junge K (2007) Psychrophilic diatoms: mechanisms for survival in freeze–thaw cycles. Algae and cyanobacteria in extreme environments. Springer, New York, pp 345–364
Owttrim GW (2006) RNA helicases and abiotic stress. Nucleic Acids Res 34:3220–3230
Owttrim GW (2013) RNA helicases: diverse roles in prokaryotic response to abiotic stress. RNA Biol 10:96–110
Pandiani F, Brillard J, Bornard I, Michaud C, Chamot S, Broussolle V (2010) Differential involvement of the five RNA helicases in adaptation of Bacillus cereus ATCC 14579 to low growth temperatures. Appl Environ Microbiol 76:6692–6697
Prochnik SE, Umen J, Nedelcu AM, Hallmann A, Miller SM, Nishii I et al (2010) Genomic analysis of organismal complexity in the multicellular green alga Volvox carteri. Science 329(5988):223–226
Provasoli L (1968) Media and prospects for the cultivation of marine algae. In: Watanabe A, Hattori R (eds) Culture and collections of algae. Proc. US-Japan Conference, Hakone
Schade B, Jansen G, Whiteway M, Entian KD, Thomas DY (2004) Cold adaptation in budding yeast. Mol Biol Cell 15:5492–5502
Schulz MH, Zerbino DR, Vingron M, Birney E (2012) Oases: robust de novo RNA-seq assembly across the dynamic range of expression levels. Bioinformatics 28:1086–1092
Thomsen MCF, Nielsen M (2012) Seq2Logo: a method for construction and visualization of amino acid binding motifs and sequence profiles including sequence weighting, pseudo counts and two-sided representation of amino acid enrichment and depletion. Nucleic Acids Res 40(1):281–287
Tripurani SK, Nakaminami K, Thompson KB (2011) Spatial and temporal expression of cold-responsive DEAD-box RNA helicases reveals their functional roles during embryogenesis in Arabidopsis thaliana. Plant Mol Biol Rep 29(4):761–768
Tuteja N, Gill SS, Tuteja R (2012) Helicases in Improving Abiotic Stress Tolerance in Crop Plants. In: Tuteja N et al (eds) Improving Crop Resistance to Abiotic Stress, vol 1. Wiley-Blackwell, Wiley-VCH Verlag GmbH and Co., Germany, pp 435–449
Umate P, Tuteja R, Tuteja N (2010) Genome-wide analysis of helicase gene family from rice and Arabidopsis: a comparison with yeast and human. Plant Mol Biol 73:449–465
Valledor L, Furuhashi T, Hanak AM, Weckwerth W (2013) Systemic cold stress adaptation of Chlamydomonas reinhardtii. Mol Cell Proteomics 12(8):2032–2047
Vashisht AA, Tuteja N (2006) Stress responsive DEAD-box helicases: a new pathway to engineer plant stress tolerance. J Photochem Photobio B 84(2):150–160
Wu G, Liu C, Liu S, Cong B, Huang X (2010) High-quality RNA preparation for cDNA library construction of the Antarctic sea–ice alga Chlamydomonas sp. ICE-L. J Appl Phycol 22(6):779–783
Yang Y, Sun Z, Ding C et al (2014) A DEAD-box RNA helicase produces two forms of transcript that differentially respond to cold stress in a cryophyte (Chorispora bungeana). Planta 240(2):369–380
Acknowledgments
This work was financially supported by a Grant of National Natural Science Foundation (41276203) for Chenlin Liu.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by L. Huang.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Liu, C., Huang, X. Transcriptome-wide analysis of DEAD-box RNA helicase gene family in an Antarctic psychrophilic alga Chlamydomonas sp. ICE-L. Extremophiles 19, 921–931 (2015). https://doi.org/10.1007/s00792-015-0768-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00792-015-0768-8