Abstract
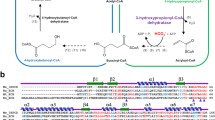
We identified the non-phosphorylated l-rhamnose metabolic pathway (Rha_NMP) genes that are homologous to those in the thermoacidophilic archaeon Thermoplasma acidophilum in the genome of the thermoacidophilic bacterium Sulfobacillus thermosulfidooxidans. However, unlike previously known 2-keto-3-deoxy-l-rhamnonate (l-KDR) dehydrogenase (KDRDH) which belongs to the short chain dehydrogenase/reductase superfamily, the putative KDRDHs in S. thermosulfidooxidans (Sulth_3557) and T. acidophilum (Ta0749) belong to the medium chain dehydrogenase/reductase (MDR) superfamily. We demonstrated that Sulth_3559 and Sulth_3557 proteins from S. thermosulfidooxidans function as l-rhamnose dehydrogenase and KDRDH, respectively. Sulth_3557 protein is an NAD+-specific KDRDH with optimal temperature and pH of 50 °C and 9.5, respectively. The K m and V max values for l-KDR were 2.0 mM and 12.8 U/mg, respectively. Sulth_3557 also showed weak 2,3-butanediol dehydrogenase activity. Phylogenetic analysis suggests that Sulth_3557 and its homologs form a new subfamily in the MDR superfamily. The results shown in this study imply that thermoacidophilic archaea metabolize l-rhamnose to pyruvate and l-lactate by using the MDR-family KDRDH similarly to that of the thermoacidophilic bacterium S. thermosulfidooxidans.




Similar content being viewed by others
References
Auernik KS, Cooper CR, Kelly RM (2008) Life in hot acid: pathway analyses in extremely thermoacidophilic archaea. Curr Opin Biotechnol 19:445–453
Baldomà L, Aguilar J (1987) Involvement of lactaldehyde dehydrogenase in several metabolic pathways of Escherichia coli K12. J Biol Chem 262:13991–13996
Brouns SJ, Turnbull AP, Willemen HL, Akerboom J, van der Oost J (2007) Crystal structure and biochemical properties of the d-arabinose dehydrogenase from Sulfolobus solfataricus. J Mol Biol 371:1249–1260
Egan SM, Schleif RF (1993) A regulatory cascade in the induction of rhaBAD. J Mol Biol 234:87–98
Eklund H, Ramaswamy S (2008) Medium- and short-chain dehydrogenase/reductase gene and protein families: three-dimensional structures of MDR alcohol dehydrogenases. Cell Mol Life Sci 65:3907–3917
Gao J, Yang HH, Feng XH, Li S, Xu H (2012) A 2,3-butanediol dehydrogenase from Paenibacillus polymyxa ZJ-9 for mainly producing R, R-2,3-butanediol: purification, characterization and cloning. J Basic Microbiol 52:1–9
Golovacheva RS, Karavaik GI (1978) A new genus of thermophilic spore-forming bacteria, Sulfobacillus. Microbiol English Translation Mikrobiologiia 47:658–664
González E, Fernández MR, Larroy C, Solà L, Pericàs MA, Parés X, Biosca JA (2000) Characterization of a (2R,3R)-2,3-butanediol dehydrogenase as the Saccharomyces cerevisiae YAL060W gene product. Disruption and induction of the gene. J Biol Chem 275:35876–35885
Hacking AJ, Aguilar J, Lin EC (1978) Evolution of propanediol utilization in Escherichia coli: mutant with improved substrate-scavenging power. J Bacteriol 136:522–530
Huff E, Rudney H (1959) The enzymatic oxidation of 1,2-propanediol phosphate to acetol phosphate. J Biol Chem 234:1060–1064
Itoh T, Suzuki K, Nakase T (2002) Vulcanisaeta distributa gen. nov., sp. nov., and Vulcanisaeta souniana sp. nov., hyperthermophilic, rod-shaped crenarchaeotes isolated from hot springs in Japan. Int J Syst Evol Microbiol 52:1097–1104
Itoh T, Suzuki K, Sanchez PC, Nakase T (1999) Caldivirga maquilingensis gen. nov., sp. nov., a new genus of rod-shaped crenarchaeote isolated from a hot spring in the Philippines. Int J Syst Bacteriol 49:1157–1163
Jung JH, Lee SB (2005) Identification and characterization of Thermoplasma acidophilum 2-keto-deoxy-d-gluconate kinase: a new class of sugar kinases. Biotechnol Bioproc Eng 10:535–539
Jung JH, Lee SB (2006) Identification and characterization of Thermoplasma acidophilum glyceraldehyde dehydrogenase: a new class of NADP+-specific aldehyde dehydrogenase. Biochem J 397:131–138
Kim S, Lee SB (2005) Identification and characterization of Sulfolobus solfataricus d-gluconate dehydratase: a key enzyme in the non-phosphorylated Entner-Doudoroff pathway. Biochem J 387:271–280
Kim S, Lee SB (2006a) Characterization of Sulfolobus solfataricus 2-keto-3-deoxy-d-gluconate kinase in the modified Entner-Doudoroff pathway. Biosci Biotechnol Biochem 70:1308–1316
Kim S, Lee SB (2006b) Rare codon clusters at 5′-end influence heterologous expression of archaeal gene in Escherichia coli. Protein Expr Purif 50:49–57
Kim SM, Paek KH, Lee SB (2012) Characterization of NADP+-specific l-rhamnose dehydrogenase from the thermoacidophilic Archaeon Thermoplasma acidophilum. Extremophiles 16:447–454
Knoll M, Pleiss J (2008) The medium-chain dehydrogenase/reductase engineering database: a systematic analysis of a diverse protein family to understand sequence-structure-function relationship. Protein Sci 17:1689–1697
Moore S, Link KP (1940) Carbohydrate Characterization I. The oxidation of aldoses by hypoiodite in methanol, II. The identification of seven aldo-monosaccharides as benzimidazole derivatives. J Biol Chem 133:293–311
Noh M, Jung JH, Lee SB (2006) Purification and characterization of glycerate kinase from the thermoacidophilic archaeon Thermoplasma acidophilum: an enzyme belonging to the second glycerate kinase family. Biotechnol Bioproc Eng 11:344–350
Petsko GA, Ringe D (2004) Protein structure and function. New Science Press, London, p 136
Rakus JF, Fedorov AA, Fedorov EV, Glasner ME, Hubbard BK, Delli JD, Babbitt PC, Almo SC, Gerlt JA (2008) Evolution of enzymatic activities in the enolase superfamily: l-rhamnonate dehydratase. Biochemistry 47:9944–9954
Sambrook J, Russel DW (2001) Molecular cloning: a laboratory manual, 3rd edn. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, New York
Segerer A, Langworthy TA, Stetter KO (1988) Thermoplasma acidophilum and Thermoplasma volcanium sp. nov. from solfatara fields. Syst Appl Microbiol 10:161–171
Sharma A, Kawarabayasi Y, Satyanarayana T (2012) Acidophilic bacteria and archaea: acid stable biocatalysts and their potential applications. Extremophiles 16:1–19
Tamura K, Stecher G, Peterson D, Filipski A, Kumar S (2013) MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol 30:2725–2729
Tian W, Skolnick J (2003) How well is enzyme function conserved as a function of pairwise sequence identity? J Mol Biol 333:863–882
Watanabe S, Makino K (2009) Novel modified version of nonphosphorylated sugar metabolism-an alternative l-rhamnose pathway of Sphingomonas sp. FEBS J 276:1554–1567
Watanabe S, Saimura M, Makino K (2008) Eukaryotic and bacterial gene clusters related to an alternative pathway of non-phosphorylated l-rhamnose metabolism. J Biol Chem 283:20372–20382
Wilson CA, Kreychman J, Gerstein M (2000) Assessing annotation transfer for genomics: quantifying the relations between protein sequence, structure and function through traditional and probabilistic scores. J Mol Biol 297:233–249
Yew WS, Fedorov AA, Fedorov EV, Rakus JF, Pierce RW, Almo SC, Gerlt JA (2006) Evolution of enzymatic activities in the enolase superfamily: L-fuconate dehydratase from Xanthomonas campestris. Biochemistry 45:14582–14597
Yelton AP, Comolli LR, Justice NB, Castelle C, Denef VJ, Thomas BC, Banfield JF (2013) Comparative genomics in acid mine drainage biofilm communities reveals metabolic and structural differentiation of co-occurring archaea. BMC Genom 14:485
Yu B, Sun J, Bommareddy RR, Song L, Zeng AP (2011) Novel (2R,3R)-2,3-butanediol dehydrogenase from potential industrial strain Paenibacillus polymyxa ATCC 12321. Appl Environ Microbiol 77:4230–4233
Acknowledgments
This work was supported by the Marine Biotechnology Program of Korean Ministry of Land, Transport and Maritime Affairs and the BK21 Program of Korean Ministry of Education.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by A. Oren.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Bae, J., Kim, S.M. & Lee, S.B. Identification and characterization of 2-keto-3-deoxy-l-rhamnonate dehydrogenase belonging to the MDR superfamily from the thermoacidophilic bacterium Sulfobacillus thermosulfidooxidans: implications to l-rhamnose metabolism in archaea. Extremophiles 19, 469–478 (2015). https://doi.org/10.1007/s00792-015-0731-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00792-015-0731-8