Abstract
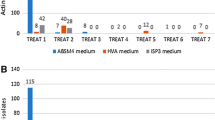
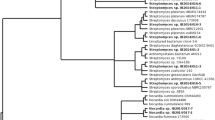
Thirty-eight actinomycetes were isolated from sediment collected from the Mariana Trench (10,898 m) using marine agar and media selective for actinomycetes, notably raffinose-histidine agar. The isolates were assigned to the class Actinobacteria using primers specific for members of this taxon. The phylogenetic analysis based on 16S rRNA gene sequencing showed that the isolates belonged to the genera Dermacoccus, Kocuria, Micromonospora, Streptomyces, Tsukamurella and Williamsia. All of the isolates were screened for genes encoding nonribosomal peptide and polyketide synthetases. Nonribosomal peptide synthetase sequences were detected in more than half of the isolates and polyketide synthases type I (PKS-I) were identified in five out of 38 strains. The Streptomyces isolates produced several unusual secondary metabolites, including a PKS-I associated product. In initial testing for piezotolerance, the Dermacoccus strain MT1.1 grew at elevated hydrostatic pressures.


Similar content being viewed by others
References
Akimoto K, Hattori M, Uematsu K, Kato C (2001) The deepest living foraminifera, Challenger Deep, Mariana Trench. Mar Micropaleontol 42:95–97
Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ (1990) Basic local alignment search tool. J Mol Biol 215:403–410
Attwell RW, Colwell RR (1984) Thermoactinomycetes as terrestrial indicators for marine and estuarine waters. In: Ortiz-Ortiz L, Bojalil LF, Yakoleff V (eds) Biological, biochemical and biomedical aspects of actinomycetes. Academic Press, Orlando, pp 441–452
Ayuso-Sacido A, Genilloud O (2005) New PCR primers for the screening of NRPS and PKS-I systems in actinomycetes: detection and distribution of these biosynthetic gene sequences in major taxonomic groups. Microb Ecol 49:10–24
Brandão FFB, Bull AT (2003) Nitrile hydrolyzing activities of deep-sea and terrestrial mycolate actinomycetes. Antonie van Leeuwenhoek 84:89–98
Brandão PFB, Clapp JP, Bull AT (2002) Discrimination and taxonomy of geographically diverse strains of nitrile-metabolizing actinomycetes using chemometric and molecular sequencing techniques. Environ Microbiol 4:262–276
Bull AT (2004) Bountiful oceans: prospecting marine microbial diversity. Screen Trends Drug Discov 5:14–16
Bull AT, Goodfellow M, Slater JH (1992) Biodiversity as a source of innovation in biotechnology. Annu Rev Microbiol 42:219–257
Bull AT, Stach JEM, Ward AC, Goodfellow M (2005) Marine actinobacteria: perspectives, challenges and future directions. Antonie van Leeuwenhoek 87:65–79
Colquhoun JA, Mexson J, Goodfellow M, Ward AC, Horikoshi K, Bull AT (1998) Novel rhodococci and other mycolate actinomycetes from the deep sea. Antonie van Leeuwenhoek 74:27–40
Courtois S, Cappellano CM, Ball M, Francou FX, Normand P, Helynck G, Martinez A, Kolvek SJ, Hopke J, Osburne MS, August PR, Nalin R, Guerineau M, Jeannin P, Simonet P, Pernodet JL (2003) Recombinant environmental libraries provide access to microbial diversity for drug discovery from natural products. Appl Environ Microbiol 69:49–55
Cross T (1968) Thermophilic actinomycetes. J Appl Bacteriol 31:36–53
Duangmal K, Ward AC, Goodfellow M (2005) Selective isolation of members of the Streptomyces violaceoruber clade from soil. FEMS Microbiol Lett 245:321–327
Felsenstein J (1985) Confidence limits on phylogeny: an appropriate use of the bootstrap. Evolution 39:783–791
Felsenstein J (1989) PHYLIP—phylogeny inference package (Version 3.2). Cladistics 5:164–166
Fiedler H-P, Bruntner C, Bull AT, Ward AC, Goodfellow M, Mihm G (2005) Marine actinomycetes as a source of novel secondary metabolites. Antonie van Leeuwenhoek 87:37–42
Finking R, Marahiel AM (2004) Biosynthesis of nonribosomal peptides. Annu Rev Microbiol 28:453–488
Ginolhac A, Jarrin C, Gillet B, Robe P, Pujic P, Tuphile K, Bertrand H, Vogel TM, Perriere G, Simonet P, Nalin R (2004) Phylogenetic analysis of polyketide synthase I domains from soil metagenomic libraries allows selection of promising clones. Appl Environ Microbiol 70:5522–5527
Goodfellow M, Haynes JA (1984) Actinomycetes in marine sediments. In: Ortiz–Ortiz L, Bojalil LF, Yakoleff V (eds) Biological, biochemical and biomedical aspects of actinomycetes. Academic Press, Orlando, pp 453–472
Gvirtzman Z, Stern RJ (2004) Bathymetry of Mariana trench-arc system and formation of the Challenger Deep as a consequence of weak plate coupling. Tectonics 23(Art no. TC2011)
Han SK, Nedashkovskaya OI, Mikhailov V, Kim SB, Bae KS (2003) Salinibacterium amurskyense gen. nov., sp. nov., a novel genus of the family Microbacteriaceae from the marine environment. Int J Syst Evol Microbiol 53:2061–2066
Hayakawa M, Nonomura H (1987) Humic acid vitamin agar, a new medium for the selective isolation of soil actinomycetes. J Ferment Technol 65:501–509
Heald SC, Brandão PFB, Hardicre R, Bull AT (2001) Physiology, biochemistry and taxonomy of deep-sea nitrile metabolising Rhodococcus strains. Antonie van Leeuwenhoek 80:169–183
Janssen PH, Yates PS, Grinton BE, Taylor PM, Sait M (2002) Improved culturability of soil bacteria and isolation in pure culture of novel members of the divisions Acidobacteria, Actinobacteria, Proteobacteria, and Verrucomicrobia. Appl Environ Microbiol 68:2391–2396
Jeanmougin F, Thompson JD, Gouy M, et al (1998) Multiple sequence alignment with CLUSTAL X. Trends Biochem Sci 23(10):403–405
Jensen PR, Dwight R, Fenical W (1991) Distribution of actinomycetes in near-shore tropical marine sediments. Appl Environ Microbiol 57:1102–1108
Jensen PR, Mincer TJ, Williams PG, Fenical W (2005) Marine actinomycete diversity and natural product discovery. Antonie van Leeuwenhoek 87:43–48
Joseph SJ, Hugenholtz P, Sangwan P, Osbone CA, Janssen PH (2003) Laboratory cultivation of widespread and previously uncultured soil bacteria. Appl Environ Microbiol 69:7210–7215
Jukes TH, Cantor CR (1969) Evolution of protein molecules. In: Munro HN (ed) Mammalian protein metabolism, vol 3. Academic Press, N Y, pp 21–132
Kato C, Li L, Nogi Y, Nakamura Y, Tamaoka J, Horikoshi K (1998) Extremely barophilic bacteria isolated from the Mariana Trench, Challenger Deep, at a depth of 11,000 meters. Appl Environ Microbiol 64:1510–1513
Kato C, Li L, Tamaoka J, Horikoshi K (1997) Molecular analyses of the sediment of the 11,000-m-deep of the Mariana Trench. Extremophiles 1:117–123
Kelly KL (1958) Centroid notations for the revised ISCC-NBS color name blocks. J Res Nat Bureau Standards USA 61:472
Ketela MM, Virpi S, Halo L, Hautala A, Hakala J, Mantsala P, Ylihonko K (1999) An efficient approach for screening minimal PKS genes from Streptomyces. FEMS Microbiol Lett 180:1–6
Ketela MM, Halo L, Manukka E, Hakala J, Mantsala P, Ylihonko K (2002) Molecular evolution of aromatic polyketides and comparative sequence analysis of polyketide ketosynthase and 16S ribosomal DNA genes from various Streptomyces species. Appl Environ Microbiol 68:4472–4479
Kim SB, Falconer C, Williams E, Goodfellow M (1998) Streptomyces thermocarboxydovorans sp. nov. and Streptomyces thermocarboxydus sp. nov., two moderately thermophilic carboxydotrophic species from soil. Int J Syst Bacteriol 48:59–68
Kyo M, Miyazaki E, Tsukioka S, Ochi H, Amitani Y, Tsuchiya T, Aoki T, Takagawa S (1995) The sea trial of “KAIKO”, the full ocean depth research ROV. Oceans 95:1991–1996
Lanoot B, Vancanneyt M, Dawyndt P, Cnockaert M, Zhang J, Huang Y, Liu Z, Swings J (2004) BOX-PCR fingerprinting as a powerful tool to reveal synonymous names in the genus Streptomyces. Emended descriptions are proposed for the species Streptomyces cinereorectus, S. fradiae, S. tricolor, S. colombiensis, S. filamentosus, S. vinaceus and S. phaeopurpureus. Syst Appl Microbiol 27:84–92
Liu W, Ahlert J, Gao Q, Pienkowski EW, Shen B, Thorson JS (2003) Rapid PCR amplification of minimal enediyne polyketide synthase cassettes leads to a predictive familial classification model. Proc Natl Acad Sci 100:11959–11963
Maldonado LA, Stach JEM, Pathom-aree W, Ward AC, Bull AT, Goodfellow M (2005) Diversity of cultivable actinobacteria in geographically widespread marine sediments. Antonie van Leeuwenhoek 87:11–18
Mincer TJ, Jensen PR, Kauffmann CA, Fenical W (2002) Widespread and persistent populations of a major new marine actinomycete taxon in ocean sediments. Appl Environ Microbiol 68:5005–5011
National Bureau of Standards (1964) ISCC-NBS color-name charts illustrated with centroid colors (supplement to national bureau of standards USA circular 553). National Bureau of Standards, USA
Olive DM, Bean P (1999) Principles and applications of methods for DNA-based typing of microbial organisms. J Clin Microbiol 37:1661–1669
Pearson K (1926) On the coefficient of racial likeness. Biometrika 18:105–117
Sadowsky MJ, Kinnel LL, Bowers JH, Schottel JL (1996) Use of repetitive intergenic DNA sequences to classify pathogenic and disease-suppressive Streptomyces strains. Appl Environ Microbiol 62:3489–3493
Sait M, Hugenholtz P, Janssen PH (2002) Cultivation of globally distributed soil bacteria from phylogenetic lineages previously only detected in cultivation-independent surveys. Environ Microbiol 4:654–666
Saitou N, Nei M (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4:406–425
Schaal KP (1977) Nocardia, Actinomadura and Streptomyces. In: CRC handbook series in clinical laboratory science, section E, clinical microbiology. CRC Press, Cleveland, OH, pp 131–158
Shirling EB, Gottlieb D (1966) Methods for characterization of Streptomyces species. Int J Syst Bacteriol 16:313–340
Stach JEM, Maldonado LA, Ward AC, Goodfellow M, Bull AT (2003a) Statistical approaches to estimating bacterial diversity in marine sediments. Appl Environ Microbiol 69:6189–6200
Stach JEM, Maldonado LA, Ward AC, Goodfellow M, Bull AT (2003b) New primers for the class Actinobacteria: application to marine and terrestrial environments. Environ Microbiol 5:828–841
Taira K, Kitagawa S, Yamashiro T, Yanagimoto D (2004) Deep and bottom currents in the Challenger Deep, Mariana trench, measured with super-deep current meters. J Oceanogr 60:919–926
Takami H, Inoue A, Fuji F, Horikoshi K (1997) Microbial flora in the deepest sea mud of the Mariana Trench. FEMS Microbiol Lett 152:279–285
Takizawa M, Colwell ER, Hill RT (1993) Isolation and diversity of actinomycetes in Chesapeake Bay. Appl Environ Microbiol 59:997–1002
Van der Peer Y, de Wachter R (1994) TREECON for Windows: a software package for the construction and drawing of evolutionary trees for the Microsoft Windows environment. Comput Appl Biosci 10:569–570
Versalovic J, Schneider M, de Bruijn FJ, Lupski JR (1994) Genomic fingerprinting of bacteria using repetitive sequence-based polymerase chain reaction. Methods Mol Cell Biol 5:25–40
Vickers JC, Williams ST, Ross GW (1984) A taxonomic approach to selective isolation of streptomycetes from soil. In: Ortiz–Ortiz L, Bojalil LF, Yakoleff V (eds) Biological, biochemical and biomedical aspects of actinomycetes. Academic Press, Orlando, pp 553–561
Ward JH (1963) Hierarchical grouping to optimise an objective function. J Am Statist Assoc 58:236–244
Weyland H (1981) Distribution of actinomycetes on the sea floor. Actinomycetes. Zbl Bakt 11(Suppl):185–193
Yamamura H, Hayakawa M, Nakagawa Y, Iimura Y (2004) Characterization of Nocardia asteroides isolates from different ecological habitats on the basis of repetitive extragenic palindromic-PCR fingerprinting. Appl Environ Microbiol 70:3149–3151
Acknowledgements
We thank the Kaiko operation team and the crew of M.S. Yokosuka for collecting sediment samples and Professor David Manning (School of Civil Engineering and Geomatics, University of Newcastle) for the use of the pressure equipment. Wasu Pathom-aree is also grateful to the Royal Thai Government and to the DPST program for financial support. ATB, MG and ACW also gratefully acknowledge support from the UK Natural Environmental Research Council (grants NER/T/S/2000/00614 and NER/T/S/ 2000/00616). We thank two anonymous reviewers for their constructive comments.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by A. Driessen
Rights and permissions
About this article
Cite this article
Pathom-aree, W., Stach, J.E.M., Ward, A.C. et al. Diversity of actinomycetes isolated from Challenger Deep sediment (10,898 m) from the Mariana Trench. Extremophiles 10, 181–189 (2006). https://doi.org/10.1007/s00792-005-0482-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00792-005-0482-z