Abstract
Previous studies identified lysine- and tryptophan-rich sequences within various cationic antimicrobial peptides. In the present study, we synthesized a series of peptides composed of lysine (K)-tryptophan (W) repeats (KW) n (where n equals 2, 3, 4 or 5) with amidation of the C-terminal to increase cationicity. We found that increases in chain length up to (KW)4 enhanced the peptides’ antibacterial activity; (KW)5 exhibited somewhat less bactericidal activity than (KW)4. Cytotoxicity, measured as lysis of human red blood cells, also increased with increasing chain length. With (KW)5, reduced antibacterial activity and increased cytotoxicity correlated with greater hydrophobicity and self-aggregation in the aqueous environment. The peptides acted by inducing rapid collapse of the bacterial transmembrane potential and induction of membrane permeability. The mode of interaction of the peptides and the phosphate groups of lipopolysaccharide was dependent upon the peptides’ ability to permeate the membrane. Longer peptides [(KW)4 and (KW)5] but not shorter peptides [(KW)2 and (KW)3] strongly bound and partially inserted into negatively charged, anionic lipid bilayers. These longer peptides also induced membrane permeabilization and aggregation of lipid vesicles. The peptides had a disordered structure in aqueous solution, and only (KW)4 and (KW)5 displayed a folded conformation on lipid membranes. Moreover, (KW)4 destroyed and agglutinated bacterial cells, demonstrating its potential as an antimicrobial agent. Collectively, the results show (KW)4 to be the most efficacious peptide in the (KW) n series, exhibiting strong antibacterial activity with little cytotoxicity.
Similar content being viewed by others
Introduction
Antimicrobial peptides (AMPs) are small proteins (~40 amino acids residues) (Brown and Hancock 2006) that can be used as potent weapons against infectious microbes, including multidrug-resistant bacteria (Hancock 2001). Indeed, AMPs constitute key parts of the innate immune systems of numerous organisms, including amphibians, and mammals. Because conventional antibiotics have specific intracellular targets, bacteria are able to develop compensatory mechanisms that render them resistant (Alanis 2005; Tenover 2006). In contrast, AMPs bind to the bacterial cell membrane and perturb its structure. This modus operandi makes AMPs impervious to bacterial resistance, since developing resistance would require microbes to alter their entire membrane lipid composition. Consequently, AMPs are attractive therapeutic candidates for protection against multidrug-resistant microbes (Fjell et al. 2011; Brogden and Brogden 2011; Giuliani and Rinaldi 2011).
AMPs adopt both α-helical and β-sheet secondary structures within hydrophobic environments (Powers and Hancock 2003). Despite differences in their secondary structures, most antimicrobial peptides share certain features, including cationicity, hydrophobicity and amphipathicity, which are essential for disruption of bacterial membranes. The cationicity of AMPs is attributable to the presence of numerous Arg and Lys residues, while amphipathicity reflects the presence of polar amino acids and non-polar, hydrophobic residues on opposite sides of the peptide secondary structure (Epand et al. 2003; Gopal et al. 2009; Kiyota et al. 1996; Song et al. 2005). The initial interaction between the bacterial membrane and an AMP is electrostatic in nature, since the bacterial membrane consists of anionic lipids such as phosphatidylglycerol and cardiolipin as well as the zwitterionic lipid phosphatidylethanolamine (Zilberstein et al. 1979; Lugtenberg and Van Alphen 1983). Once the polar side of the AMP, which is enriched with charged residues, binds to the anionic bacterial membrane, the hydrophobic region of the peptide attaches to the hydrophobic core of the lipid bilayer, thereby causing a partition.
Although charge and amphipathicity greatly influence the activity of AMPs, naturally occuring AMPs also differ greatly in length, making it very difficult to correlate chain length and antimicrobial activity. Because knowing the optimal length of AMPs could facilitate de novo design of AMPs (Deslouches et al. 2005), we designed and synthesized a series of peptides of different length, and then determined their modes of action using lipid bilayer and bacterial membrane models. We found that the length of the peptide chain greatly influences target selection, permeabilization of the bacterial membrane, and agglutination of bacterial cells.
Materials and methods
Materials
Rink amide 4-methylbenzhydrylamine resin, fluoren-9-ylmethoxycarbonyl (Fmoc) amino acids and other reagents for peptide synthesis were purchased from Calbiochem-Novabiochem (La Jolla, CA, USA). Acrylamide was from Sigma-Aldrich (St. Louis, MO, USA). Cholesterol (CH, from porcine liver), l-α-phosphatidylethanolamine (PE, from E. coli), egg yolk l-α-phosphatidylcholine (PC) and l-α-phosphatidylglycerol (PG, from E. coli) were from Avanti Polar Lipids Co. (Alabaster, AL, USA). Calcein, 5-carboxytetramethylrhodamine (TAMRA), 3,3′-diethylthiodicarbocyanine iodide (DiSC3-5) and SYTOX Green were from Molecular Probes (Eugene, OR, USA). All other reagents were of analytical grade. Buffers were prepared using double distilled water (Millipore Co.).
Bacterial strains
Escherichia coli (KCTC 1682), Salmonella typhimurium (KCTC 1926), Pseudomonas aeruginosa (KCTC 1637), Staphylococcus aureus (KCTC 1621), Bacillus subtilis (KCTC 1918), and Listeria monocytogenes (KCTC 3710) were obtained from the Korean Collection for Type Cultures (KCTC). Drug-resistant E. coli strains (CCARM 1229 and CCARM 1238), S. typhimurium strains (CCARM 8007, CCARM 8009, and CCARM 8013), and S. aureus strains (CCARM 3089, CCARM 3090, CCARM 3108, CCARM 3114, and CCARM 3126) were from the Culture Collection of Antibiotic-Resistant Microbes (CCARM) at Seoul Women’s University, Korea.
Peptide synthesis and purification
The peptides KWKW-NH2 [(KW)2], KWKWKW-NH2 [(KW)3], KWKWKWKW-NH2 [(KW)4], and KWKWKWKWKW-NH2 [(KW)5] were synthesized using the solid-phase method with Fmoc chemistry on a solid support of rink amide 4-methylbenzhydrydrylamine resin. Thereafter, 0.1 M N-hydroxy benzotriazole (HOBt), 0.45 M 2-(1H-benzotriazole-1-yil)-1,1,3,3-tetramethyluroniumhexafluorophosphate (HBTU) in dimethylformamide (DMF) and 2 M N,N-diisopropyl ethylamine (DIEA) in N-methylpyrrolidone (NMP) were used as coupling reagents, and a tenfold excess of Fmoc-amino acid was added during every coupling cycle. After a final deprotection step using a solution of 20 % piperidine in DMF, cleavage was carried out using a mixture of TFA/water/triisopropylsilane (90:5:5) for 2 h at room temperature. The crude peptides were repeatedly extracted with diethyl ether and then purified using reverse phase preparative HPLC on a Vydac C18 column (4.6 × 250 mm, 300 Å, 5 μm). The molecular masses of the peptides were confirmed using a matrix-assisted laser desorption ionization mass spectrometer (MALDI II, Kratos Analytical Ins.).
Antibacterial activity
The antibacterial activities of the peptides against Gram-negative, Gram-positive and antibiotic-resistant bacterial strains were evaluated using the microbroth dilution method (Park et al. 2003). Aliquots of culture medium containing bacterial suspensions in mid-logarithmic phase at a concentration of 2 × 105 colony forming units (CFU)/ml were added to each well of a multiwell plate containing peptide solution serially diluted 2× in 10 mM sodium phosphate buffer (pH 7.2) or phosphate-buffered saline (PBS, 1.5 mM KH2PO4, 2.7 mM KCl, 8.1 mM Na2HPO4, 150 mM NaCl, pH 7.2). Growth inhibition was evaluated by measuring the absorbance at 620 nm using a Versa-Max microplate Elisa Reader (Molecular Devices Co., Sunnyvale, CA, USA) after 24 h of incubation at 37° C. The minimum inhibitory concentration (MIC) was defined as the minimal peptide concentration that completely inhibited bacterial growth. All MIC measurements were the average of 3–4 independent experiments.
Human red blood cell (hRBC) hemolysis
Hemolytic activities of the peptides were assessed using heparinized hRBCs collected from healthy donors. Fresh hRBCs were washed three times by centrifugation at 800×g for 10 min in PBS (pH 7.2) and then resuspended in PBS. The peptides were then dissolved in PBS and added to 100 μl of stock hRBCs suspended in PBS (final RBC concentration, 4 % v/v), after which the samples were incubated with gentle agitation for 60 min at 37° C before centrifugation for 10 min at 800×g. Absorbance of the supernatants was recorded at 414 nm. In addition, control samples for zero hemolysis and 100 % hemolysis were incubated with PBS (A blank) or 0.1 % Triton X-100 (A triton), respectively. Melittin was used as a reference hemolytic peptide. Percent hemolysis was calculated according to the following equation:
Each measurement was conducted in triplicate (Kim et al. 2011).
Computational modeling
The structures of the peptide models were built using ChemOffice Desktop 2004 for Windows (CambridgeSoft (CS) Corporation, MA, USA). The structures of linear peptides were generated and the energies minimized using Chem3D and the molecular mechanics method (MM2) (Burkert and Allinger 1982). The sequences of the peptides were initially added using standard ChemDraw Ultra, after which the N-and C-termini of the peptide groups were charged by changing the balances. Amidation of the C-terminus of each peptide was carried out using Chem3D. Energy minimizations were run for all of the peptides using the Chem3D and MM2 methods. Finally, optimized peptides were imported into the Pymol program for visualization and figure generation (DeLano 2002).
Aggregation states of peptides in aqueous solution
We compared the aggregation states of peptides in water and PBS by examining Trp fluorescence as a function of peptide concentration from 1 to 18 μM. Changes in emission were recorded using a spectrofluorometer (Perkin-Elmer LS55, Mid Glamorgan, UK) at an excitation wavelength of 280 nm and an emission wavelength of 300–400 nm.
CD spectroscopy was used to determine the aggregation states of the peptides in PBS at various concentrations ranging from 25 to 150 μM. The CD spectra were recorded between 190 and 250 nm on a Jasco 810 spectropolarimeter (Jasco, Tokyo, Japan) equipped with a temperature control unit. The recordings were made using a quartz cell with a 0.1-cm path length at 25° C. At least four scans over the 190–250 nm wavelength range were conducted.
Confocal laser-scanning microscopy (CLSM)
Fluorescently labeled (KW)4 peptide was used for the CLSM study. Fluorescent labeling of the N-termini of the peptides was accomplished using TAMRA in DMF (3–4 eq.) containing 5 % (v/v) DIEA (Ghosh et al. 1997).
Escherichia coli CCARM 1229 or S. aureus CCARM 3090 cells in mid-logarithmic phase were harvested by centrifugation, washed and re-suspended in PBS (pH 7.2) to a concentration of 2 × 105 CFU/ml. TAMRA-labeled peptide was then added to 100 μl of the cell suspension to a concentration of 12.5 μM, after which the cells were incubated at 37° C for 10 min, pelleted by centrifugation at 3,000×g for 5 min, and washed three times with PBS. The action site of TAMRA-labeled (KW)4 was then examined using an inverted LSM510 laser-scanning microscope (Carl Zeiss, Göttingen, Germany).
Dansyl polymyxin B (PMB) displacement assay
The binding affinity of each peptide for lipopolysaccharide (LPS) was determined using dansyl PMB displacement assays (Moore et al. 1986). Dansyl PMB (4 μM/ml) and P. aeruginosa LPS (9 μg/ml) were mixed in 1 ml of 5 mM HEPES (pH 7.2) for optimal fluorescence. Reductions in fluorescence were used to calculate the percent displacement of dansyl PMB after peptide treatment. Buffer blank was subtracted from the peptide spectra. Spectra were recorded on a Perkin-Elmer LS-50B spectrofluorimeter using a cuvette with of 1-cm path length. The excitation and emission wavelengths were 340 and 485 nm, respectively.
Membrane depolarization
Peptide-induced membrane depolarization was examined using Gram-positive and Gram-negative bacteria as described previously (Papo et al. 2002; Sal-Man et al. 2002). Briefly, E. coli CCARM 1229 and S. aureus CCARM 3090 were grown to mid-logarithmic phase at 37° C with gentle agitation. The cells were then washed twice in buffer (20 mM glucose, 5 mM HEPES, PH 7.3) and re-suspended to an OD600 of 0.05 in similar buffer containing 0.1 M KCl. The bacterial cells were then incubated with 1 μM DiSC3-5 until the fluorescence reached a stable baseline. After addition of the indicated concentrations of peptide, changes in fluorescence were measured using an excitation wavelength of 622 nm and emission wavelength of 670 nm. The fluorescence of 0.1 % Triton X-100-treated cells served as a positive control for maximum depolarization.
SYTOX Green uptake
Cultured cells (E. coli CCARM 1229 and S. aureus CCARM 3090) were re-suspended (2 × 107 CFU/ml) in PBS and incubated with 1 μM SYTOX Green for 10 min in the dark. Peptides (12.5 μM) were then added to the cell suspension, and the fluorescence was monitored for 60 min at an excitation wavelength of 485 nm and emission wavelength of 520 nm.
Kinetic studies
The kinetics of the bactericidal activity of peptides were evaluated using E. coli CCARM 1229 and S. aureus CCARM 3090. Mid-logarithmic growth phase bacteria (2 × 105 CFU/ml) were incubated with peptides at 1× or 2× the MIC at 37° C. Aliquots were removed at the indicated times, appropriately diluted, and plated on LB agar plates. CFUs were counted after 16 h of incubation at 37° C. All measurements were carried out in triplicate.
Preparation of small and large unilamellar vesicles (SUVs and LUVs)
SUVs were prepared by sonication of the required amount of PE:PG (7:3, w/w) or PC:CH (10:1, w/w). Dry lipids were dissolved in chloroform in a glass vessel, after which the solvent was removed using a stream of nitrogen gas. Any remaining trace amounts of organic solvent were then completely removed by lyophilization overnight, and the remaining dry lipid film was resuspended in PBS at (pH 7.2) with gentle vortex mixing. The lipid suspensions were then sonicated at 40° C until clear using a bath-type ultrasonicator and extruded 14 times using an Avanti Mini-Extruder (Avanti Polar Lipids Inc., Alabaster, AL, USA) with 0.05 μm polycarbonate filters.
LUVs were prepared using the freeze–thaw method (Mayer et al. 1986). Briefly, dry lipid films were resuspended in 1–2 ml of appropriate buffer by vortexing. LUVs were prepared through nine freeze–thaw cycles in liquid nitrogen, followed by incubation in a water bath at 50° C. The suspensions were then extruded 14 times through 0.2-μm polycarbonate membranes, and lipid concentrations were determined using standard phosphate assays (Stewart 1980).
Calcein leakage from liposomes
PE:PG (7:3, w/w) and PC:CH (10:1, w/w) LUVs with entrapped calcein were prepared by vortexing the dried lipids in a dye buffer solution (70 mM calcein, PBS, pH 7.4). The resultant suspension was then subjected to nine cycles of freeze–thaw in liquid nitrogen, followed by separation of the calcein-containing vesicles from free calcein using gel filtration chromatography on a Sephadex G-50 column. LUVs with entrapped calcein in a suspension containing 2.5 μM lipid were then incubated for 25 min with various concentrations of peptide (0.03–0.25 μ). The release of fluorescent calcein was assessed using a spectrofluorometer at an excitation wavelength of 480 nm and an emission wavelength of 520 nm. Complete (100 %) release was induced by addition of 0.1 % Triton X-100; spontaneous leakage was negligible. All experiments were conducted at 25° C, and the apparent percentage of released calcein was calculated using the following equation (Matsuzaki et al. 1999):
where F and F t are the fluorescence intensity before and after the addition of detergent, respectively, and F o is the fluorescence of intact vesicles.
Liposome aggregation
The aggregation of lipid vesicles was monitored through visible absorbance measurements. Peptides (5, 10, 20, and 40 μM) in PBS (pH 7.2) were added to a suspension of 400 μM PE:PG LUVs (7:3, w/w). Absorbance was measured at 405 nm using a microplate Autoreader before and after the addition of the peptides (Oren et al. 1999). Increased absorbance indicated liposomal aggregation.
Trp fluorescence and acrylamide quenching assays
The fluorescence emission spectrum of Trp within the peptides was monitored using a spectrofluorometer in the presence of PE:PG (7:3, w/w) or PC:CH (10:1, w/w) SUVs, which were used to minimize differential light scattering effects (Mao and Wallace 1984). Each peptide was added to 1 ml of 200 μM liposomes, after which the peptides and liposomes (molar ratio 1:100) were allowed to interact for 10 min at 25° C. The fluorescence was then measured at an excitation wavelength of 280 nm and an emission wavelength from 300 to 400 nm.
Fluorescence quenching experiments were conducted using acrylamide as the quencher. The concentration of acrylamide in the cuvette ranged from 0.04 to 0.20 M. The effect of acrylamide on the fluorescence of each peptide was analyzed using the Stern–Volmer equation
where F 0 and F are the fluorescence intensities in the presence or absence of acrylamide, respectively, K SV is the Stern–Volmer quenching constant and (Q) is the concentration of acrylamide.
CD spectroscopy
The CD spectra for 50 μM peptide dissolved in PBS alone (pH 7.2), PBS containing 1 mM PE:PG (7:3, w/w) vesicles, or PBS containing 1 mM PC:CH (10:1, w/w) vesicles were scanned in the presence or absence of LPS (0.1 %) dissolved in PBS. The CD data represent the average values of three separate recordings with four scans per sample.
Scanning electron microscopy (SEM)
Mid-logarithmic phase cultured E. coli CCARM 1229 cells were re-suspended at 2 × 107 CFU/ml in PBS (pH 7.2) and incubated with 12.5 μM (KW)4 for 1 h at 37° C. The bacteria were then pelleted by centrifugation at 3000×g for 5 min and washed twice in PBS. After removing the supernatants, the pellets were fixed for 3 h at 4° C in 500 μl of 5 % (v/v) glutaraldehyde in 0.2 M sodium-cacodylate buffer (pH 7.4). The samples were then extensively washed with 0.1 M sodium-cacodylate buffer and treated with 1 % (w/v) osmium tetroxide in 0.1 M sodium-cacodylate buffer for 1 h at 4° C in the dark. Thereafter, the bacteria were washed twice with 5 % (w/v) sucrose in the same buffer, and dehydrated in a sequential 20, 40, 60, 80, 95 and 100 % ethanol series. After lyophilization and coating, the samples were examined in a scanning electron microscope (Hitachi S-2400N, Japan).
Bacterial agglutination assay
Overnight cultures of E. coli CCARM 1229, S. aureus CCARM 3090 and P. aeruginosa were washed with 10 mM sodium phosphate (pH 7.4) containing 10 % TSB media. After adjusting the concentration to 2 × 108 CFU/ml, 100 μl aliquots of the bacterial suspension were incubated with 100 μl of peptide solution (6.25–50 μM) in a polystyrene plate for 1 h at 37° C. The bacteria were then stained for 10 min with 1 % (w/v) crystal violet. After the supernatants containing non-aggregated cells were carefully removed, each well was photographed using an Olympus Inverted Microscope (Olympus 1X71, Tokyo, Japan) equipped with digital camera (Olympus DP71).
Results
Effects of hydrophobicity and length on the antibacterial and hemolytic activities of (K–W) n peptides and computer modeling of peptide structures
The observed molecular weights of the synthetic (KW) n peptides were well matched to the calculated values. To determine the antibacterial and hemolytic activities of the peptides, we used RP-HPLC to assess the level of hydrophobicity of the synthetic peptides (Table 1). We found that the retention times, and thus the relative hydrophobicity, of the (KW) n peptides were in the order (KW)2 < (KW)3 < (KW)4 < (KW)5, indicating that the hydrophobicity of the synthetic peptides increased with the addition of KW motifs.
The antibacterial activities of the peptides in PBS and low ionic strength buffer [sodium phosphate (SP) buffer] are summarized in Table 1. In general, the antibacterial activity of the peptides increased with increasing chain length up to (KW)4. Whereas (KW)2 was inactive against all strains, (KW)3 and (KW)4 exhibited activity against S. typhimurium, P. aeruginosa and L. monocytogenes. Moreover, (KW)4 exhibited stronger antibacterial activity than the other peptides against all bacterial strains. Interestingly, the antibacterial activity of (KW)5 was similar to or somewhat weaker than that of (KW)4, indicating that, with (KW)5, increasing chain length did not lead to an increase in antibacterial activity.
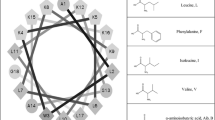
Neither (KW)2 nor (KW)3 induced hemolysis at a concentration of 200 μM (Table 1). In contrast, (KW)4 and (KW)5 caused 8 and 71 % hemolysis, respectively. The greater hemolytic activity of (KW)5 likely reflects its greater hydrophobicity. In addition, Feder et al. (2000) reported a linear correlation between hemolysis and the aggregation state. We therefore next examined the aggregation state of (KW)5 in aqueous solution. Computer modeling revealed that the clustering of KW motifs within (KW) n peptides, resulted in different spatial arrangements and surface areas (Fig. 1).
Structure and organization of (KW) n peptides in aqueous solution
To investigate why (KW)5 exhibited less antibacterial activity and greater hemolytic activity than (KW)4, we assessed the self-aggregation of (KW)4 and (KW)5 in water and PBS. As determined from the Trp fluorescence, neither peptide aggregated in water (Fig. 2a). In PBS, the Trp fluorescence maximum from (KW)5 dose dependently shifted from 352 to 340 nm due to its self-aggregation (Fig. 2b). On the other hand, the Trp fluorescence of (KW)4 did not change, implying no self-aggregation. We did not consider (KW)2 or (KW)3 in the self-aggregation assays because of their short chain lengths.
Structure and organization of (KW)4 and (KW)5 in aqueous solution. Aggregation states of the peptides in aqueous solution were determined based on Trp fluorescence and are shown as functions of the peptide concentration. The wavelength at the emission maximum was taken for plotting. (KW)4 (filled circles), (KW)5 (filled triangles): water (a) and PBS (pH 7.2) (b). Concentration-dependent CD spectroscopy was used to examine the conformations of the soluble and aggregation states of the peptides. Concentration-dependent CD spectra for (KW)4 (c) and (KW)5 (d) in PBS (pH 7.2): 25 μM (solid line), 50 μM (dashed line), 100 μM (dotted line) and 150 μM (dashed-dotted line)
We further analyzed the aggregation states using CD spectroscopy. As shown in Fig. 2c and d, the peptides in PBS had a negative band at 200 nm, which is characteristic of random coil. These peptides also had a band at 225 nm due to the Trp side chain, which was consistent with earlier reports (Liu et al. 2007; Woody 1994; Ladokhin et al. 1997). Because (KW)5 assumes a random coil conformation, it failed to form a folded conformation at concentrations of 25 and 50 μM. At higher concentrations (100 and 150 μM), however, the negative band of (KW)5 was shifted to a slightly longer wavelength, suggesting that a small fraction of the peptide adopted a folded conformation or was weakly aggregated (Fig. 2d). In contrast, (KW)4 peptide maintained its random coil conformation in PBS at all tested concentrations (Fig. 2c).
Localization of fluorescence-labeled peptides
To determine the site targeted by (KW)4 in E. coli CCARM 1229 and S. aureus CCARM 3090, the bacteria were treated with the peptide and observed under CLSM. As shown in Fig. 3, the peptide bound to the surface of E. coli and S. aureus cells (Fig. 3), confirming that these peptides interact with the bacterial cell membrane.
Confocal laser-scanning micrographs images of E. coli CCARM 1229 (a) and S. aureus CCARM 3090 (b) cells treated with TAMRA-labeled-(KW)4. The cells were treated with 12.5 μM TAMRA-(KW)4 for 10 min at 37° C in PBS (pH 7.2). From left to right: TAMRA, differential interference contrast (DIC), merged images
Interaction of peptides with LPS
We next used dansyl PMB displacement assays to determine the binding affinity of each peptide for LPS. The method is based on enhanced dansyl fluorescence upon the interaction of LPS. This fluorescence is decreased when peptides bind to LPS and displace the dansyl PMB. As shown in Fig. 4a, all four tested peptides dose-dependently bound to LPS, with (KW)5 exhibiting the highest affinity. At a concentration of 20 μM, (KW)5 displaced 81 % of dansyl PMB from LPS. By comparison, (KW)4, (KW)3 and (KW)2 displaced 70, 55 and 40 %, respectively, at the same concentration. These data clearly indicate a correlation between chain length and LPS binding affinity.
Binding affinities of the peptides for LPS, as measured using dansyl PMB displacement assays and CD spectroscopy. P. aeruginosa LPS (9 μg) was incubated with dansyl PMB (4 μM) for 5 min, after which fluorescence was measured at an excitation wavelength of 340 nm and emission wavelength of 485 nm. Peptides were added at different concentrations, and dansyl PMB fluorescence was measured after 5 min (a). CD spectra for the peptides (50 μM) were measured in the presence of 0.1 % LPS (b): (KW)2 (dashed-dotted line), (KW)3 (dotted line), (KW)4 (dashed line), and (KW)5 (solid line)
We also used the peptides’ CD spectra to predict their interactions with LPS. When the peptides were studied at 50 μM in a 0.1 % LPS suspension, (KW)2 and (KW)3 exhibited no secondary structure, while the longer peptides adopted folded conformations (Fig. 4b), which is indicative of their strong binding to LPS and was highly correlated with their displacement of dansyl PMB.
Membrane lytic and bactericidal activities of the peptides
We examined the peptides’ capacity to disrupt the cytoplasmic membrane in E. coli and S. aureus. With the exception of (KW)2, the peptides dissipated the membrane potential of intact E. coli CCARM 1229 (Fig. 5a) and S. aureus CCARM 3090 (Fig. 5b) in a dose-dependent manner, and the longer peptides were more effective than the shorter ones. Moreover, the capacity of (KW)4 and (KW)5 to disrupt the cytoplasmic membrane correlated well with their antibacterial activities against the same bacteria (Table 1). Inactive (KW)2 did not depolarize the membrane, while (KW)3 elicited less depolarization than the longer peptides.
Membrane-disruption and bactericidal activity. Dose–response curves for peptide-induced membrane depolarization in intact E. coli CCARM 1229 (a) and S. aureus CCARM 3090 (b). Membrane depolarization was monitored as an increase in the fluorescence of DiSC3-5 (excitation wavelength 620 nm, emission wavelength 670 nm) after addition of various concentrations of peptide: (KW)2 (filled diamonds), (KW)3 (filled squares), (KW)4 (filled circles), and (KW)5 (filled triangles). The increase in fluorescence obtained using 0.1 % Triton X-100 was taken as 100 %. Membrane permeabilization was monitored based on entry of SYTOX Green dye. Bacterial cells (2 × 107 CFU/ml) in PBS were incubated with 1 μM SYTOX Green dye. Peptides were added at a concentration of 12.5 μM, after which uptake of SYTOX Green through the E. coli (c) or S. aureus (d) plasma membrane was measured based on the time course of fluorescence changes at an excitation wavelength of 485 nm and emission wavelength of 520 nm: (KW)2 (filled diamonds), (KW)3 (filled squares), (KW)4 (filled circles), (KW)5 (filled triangles), and Melittin (open circles). Kinetics of the bactericidal activity against E. coli CCARM 1229 (e) and S. aureus CCARM 3090 (f). Bacteria treated with the respective peptides were diluted at the appropriate times and then plated on LB agar. CFUs were then counted after 16 h of incubation at 37° C. Black (KW)4, white (KW)5, squares and triangles 1 and 2 times the MIC, respectively; cells (2 × 105 CFU/ml) incubated in the absence of any peptide served as controls
The ability of the (KW) n peptides to permeabilize the cell membrane was determined based on the uptake of SYTOX Green, a fluorescent dye, by E. coli and S. aureus cells. After the addition of 12.5 μM peptide in PBS, influx of SYTOX Green fluorescence was measured for 60 min. Treatment with (KW)4 or (KW)5 efficiently activated uptake of the dye within 35 min in both E. coli (Fig. 5c) and S. aureus (Fig. 5d). (KW)5 induced internalization of SYTOX Green more quickly than (KW)4, which is indicative of its more efficient interaction and partition within the bacterial lipid bilayer, and is consistent with its greater hydrophobicity. (KW)3 elicited less uptake of SYTOX Green than (KW)4 or (KW)5. These results suggest that active peptides exert their antibacterial activities through induction of membrane damage.
To further investigate the bactericidal activities of (KW)4 and (KW)5, we performed a time-killing assay against E. coli CCARM 1229 and S. aureus CCARM 3090. As shown in Fig. 5e and f, at concentrations equal to or above the MIC, bacterial counts significantly declined within 35 min. (KW)5 killed bacteria more quickly than (KW)4, possibly due to its longer chain length and/or greater hydrophobicity, which is consistent with its faster permeabilization of the target bacterial membrane (Fig. 5c, d).
Calcein release and turbidity assay in liposomes
We also evaluated the abilities of these peptides to cause leakage of entrapped calcein from PE:PG (7:3, w/w) LUVs, which mimic a bacterial membrane. At a lipid-to-peptide molar (L/P) ratio of 2.5:1, (KW)4 released 81 % of entrapped calcein, while (KW)5 released about 73 %. (KW)3 exhibited moderate activity, but (KW)2 was inactive (Fig. 6a). We also examined the leakage of entrapped calcein from using zwitterionic liposomes composed of PC:CH (10:1, w/w), which mimic eukaryotic membranes. At a L/P ratio of 2.5:1, (KW)5 and (KW)4 released 80 and 30 % of entrapped calcein, respectively (Fig. 6b), while (KW)2 and (KW)3 elicited no release of calcein from zwitterionic vesicles. The peptides’ respective abilities to release dye from PC:CH vesicles is therefore consistent with their hemolytic activities (Table 1).
Peptide actions on lipid bilayers. Percent leakage of calcein from PE:PG (7:3, w/w) (a) and PC:CH (10:1, w/w) vesicles (b) at pH 7.4 was measured for 25 min after the addition of various doses of peptide. c LUV aggregation. Solutions containing various concentrations of peptide were added to a suspension of 400 μM PE:PG (7:3, w/w) LUVs, after which aggregation was monitored based on changes in the absorbance of the LUVs at 405 nm: (KW)2 (filled diamonds), (KW)3 (filled squares), (KW)4 (filled circles), and (KW)5 (filled triangles)
We then measured changes of liposome turbidity to monitor the effects of the peptides on the size of PE:PG (7:3, w/w) vesicles. We found that the peptides increased the turbidity of PE:PG (7:3, w/w) vesicles, indicating an increase in vesicle size, most likely due to vesicle lysis and aggregation (Fig. 6c) (Domingues et al. 2009).
Characterization of the Trp environment using fluorescence spectroscopy
To characterize the Trp environment of the peptides, we used fluorescence spectroscopy to examine the binding of the peptides to lipid bilayers. Trp emission spectra was recorded in the presence of negatively charged PE:PG (7:3, w/w) or zwitterionic PC:CH (10:1, w/w) vesicles or in PBS (Table 2). For all of the peptides except (KW)5, the fluorescence maximum in buffer was around 353 nm, indicating that the Trp residues were situated in a more hydrophilic environment. For (KW)5, the fluorescence spectra suggest that Trp is shifted from a polar to less polar environment, possibly due to self-aggregation of the peptide in PBS (Fig. 2).
The ability of the peptides to bind to PE:PG (7:3, w/w) vesicles was in the order: (KW)4 > (KW)5 > (KW)3 > (KW)2. In addition, (KW)4 exhibited a large blue shift, indicating that its Trp side chains partitioned preferentially into a more rigid, and hydrophobic environment within the PE:PG (7:3, w/w) vesicles. Similarly, the Trp emission from (KW)5 (342 nm) also exhibited a blue shift in the presence of PC:CH (10:1, w/w) vesicles, indicating migration of Trp residues to a more hydrophobic environment (Burstein et al. 1973). On the other hand, no blue shifts were detected for (KW)2, (KW)3 or (KW)4 in the presence of the more hydrophobic environment of PC:CH (10:1, w/w) vesicles.
We evaluated the ability of Trp residues to access the lipid bilayer by measuring the Stern–Volmer quenching constants (K SV) using soluble acrylamide as a quencher. Free Trp residues in aqueous solution were fully quenched by acrylamide (K SV = ~9 M−1) (Wimley et al. 1996), and the K SV values for (KW)2, (KW)3, (KW)4 and (KW)5 in PBS were (approximately) 15, 15, 14, and 11 M−1, respectively (Table 2). That the K SV value for (KW)4 was 1.6 M−1 in the presence of PE:PG (7:3, w/w) vesicles, indicates that the Trp residues of (KW)4 were more protected in PE:PG (7:3, w/w) vesicles than PC:CH (10:1, w/w) vesicles (Table 2). This tendency correlated well with the potent antibacterial activity of (KW)4. In contrast, the hemolytic peptide (KW)5 had a lower K SV value, indicating that its Trp residues were more anchored within the hydrophobic core of the zwitterionic phospholipids, which correlates well with the strong induction of calcein leakage from the vesicles (Fig. 6b).
Structures of the peptides in liposomes
We determined the secondary structures of the peptides in aqueous solution and lipid membranes. (KW)2 and (KW)3 displayed no secondary structure in aqueous solution (data not shown) or lipid membranes. (KW)4 and (KW)5 adopted folded conformations on liposomes (Fig. 7). (KW)5 displayed a positive band at 197 nm and a negative band at 210 nm on PC:CH vesicles (Fig. 7b), indicating that its side chain partitioned preferentially into the more rigid and hydrophobic environment of the lipid bilayer, which is consistent with its potent hemolytic activity.
Observation of (KW)4 activity in E. coli using SEM
To examine the action of (KW)4 against E. coli CCARM 1229 in more details, we used SEM to observe the morphological changes that occur upon incubation of the peptide with bacteria. In the absence of peptide, the bacteria had a smooth surface (Supplementary Fig. 1A), but in the presence of 12.5 μM peptide the treated bacteria appeared clumped with more crenated surfaces (Supplementary Fig. 1B).
Bacterial agglutination
To confirm the bacterial aggregation, we incubated E. coli, S. aureus, or P. aeruginosa with various concentrations of peptide for 1 h. At higher concentrations of peptide, there was greater agglutination and bacterial clumping (Supplementary Fig. 2). (KW)4 induced agglutination of all three bacterial strains. Bacterial aggregation was not observed in the absence of peptide. These results demonstrate the agglutination-inducing activity of (KW)4 peptide against both Gram-negative and Gram-positive bacteria.
Discussion
De novo design of short peptides is an alternative strategy for the development of AMPs that are selective for bacteria, with little or no effect on human cells. We designed peptides comprised of alternating Lys and Trp residues, which we previously showed to have antimicrobial activity (Gopal et al. 2011). When choosing a cationic amino acid for the present study, we chose Lys over Arg because, the guanidinium group in the side chain of Arg is active in both microbial and mammalian cell membranes (Yang et al. 2003). Lys also possesses one positive charge but lacks a guanidinium group, making it less toxic to mammalian cells (Andra et al. 2007). In addition, the cost of producing Arg-containing peptides is higher than that of producing Lys-containing ones, which is an important factor when considering drug design and development. Finally, Trp was selected, based on reports indicating that AMPs containing Trp are more active than those containing either Tyr or Phe (Dathe et al. 2004; Strom et al. 2003; Subbalakshmi et al. 2000). One explanation could be that the bulkier side chain of Trp may interact more strongly with the hydrophobic membrane core than other non-polar amino acids, such as Phe and Tyr (Strom et al. 2002; Wimley and White 1996).
The results obtained in earlier studies of the effect of chain length on the activity of AMPs have varied. In one study of a (RW) n series, chain length correlated well with antibacterial and hemolytic activities (Liu et al. 2007). But in another study of H-(LARL)3-(LRAL) n peptides, increasing chain length reduced the antibacterial activity while increasing hemolysis (Niidome et al. 2005). In the present study, we observed that antibacterial activity improved with increasing chain length up to (KW)4; there was no further increase in antibacterial activity when the chain length exceeded eight amino acids. The order of the activity was (KW)2 < (KW)3 < (KW)4 > (KW)5. (KW)5 exhibited nearly the same antibacterial activity as (KW)4, except when treating S. typhimurium and P. aeruginosa in PBS. The observed increase in antibacterial activity as the peptide chain lengthened is attributable to increases in the cationicity, hydrophobicity and clustering of the KW motifs (Fig. 1). When chain length exceeded the optimum eight residues, as with (KW)5, antibacterial activity declined, and there was a greater propensity for aggregation in aqueous buffer due to the increased hydrophobicity. The observed difference in the peptides’ ability to aggregate in PBS and SP buffer may be attributable to differences in the ionic strengths of the two buffers. Our results suggest that (KW)5 was soluble and weakly self-aggregated in PBS (Fig. 2).
The hemolytic activities of the (KW) n peptides increased with chain length. Short peptides were inactive, and (KW)4 showed modest hemolytic activity at high concentrations. (KW)5 exhibited marked hemolytic activity, reflecting an increase in hydrophobicity as well as the peptide’s ability to weakly aggregate. Several studies have suggested that AMPs in a self-aggregated state in aqueous solution, where there is an increase in hydrophobic interactions, mediate hemolytic activity (Feder et al. 2000; Javadpour and Barkley 1997). Our results with (KW)5 are consistent with that idea. Only (KW)4 exhibited good antibacterial activity without a high degree of hemolysis. This may reflect an optimal balance of cationicity and hydrophobicity, which are features needed for broad-spectrum antibacterial activity without hemolysis (Javadpour et al. 1996; Fernandez-Lopez et al. 2001).
AMPs kill bacteria either through permeabilization of the cytoplasmic membrane or by binding with intracellular targets like nucleic acids (Brogden 2005). Our CLSM results indicate that (KW) n peptides interact with the bacterial surface (Fig. 3), suggesting (KW) n act by damaging the cytoplasmic membrane, rather than binding to nucleic acids. Gram-negative bacteria have two-cell membranes, the outer membrane and inner cytoplasmic membrane. The outer leaflet of the outer membrane consists of LPS, making it necessary for the peptide to bind to LPS and cross the outer membrane before entering the periplasmic space (Farnaud et al. 2004; Bhattachariya and Ramamoorthy 2009). For this reason, we examined the peptides’ ability to bind LPS. We found that longer peptides strongly bound LPS (Fig. 4) and induced membrane depolarization (Fig. 5a, b). The higher affinities of (KW)4 and (KW)5 for LPS correlated with their abilities to induce membrane depolarization, suggesting they effectively disrupted the outer membrane and accessed the cytoplasmic membrane. The peptides then appeared to damage the inner membrane (Fig. 5c, d) to exert their bactericidal action (Fig. 5e, f).
Using lipid bilayers, it was previously shown that the mechanism of action of AMPs involves two stages: initial electrostatic interaction of the cationic residues (Lys or Arg) of the peptides with the anionic lipids of the bacterial membrane (Juffer et al. 2001; Shepherd et al. 2001), followed by hydrophobic interaction of aromatic side chain-containing amino acids with the lipid bilayer (Schibli et al. 2002; Wimley and White 1996). Our results are consistent with that mechanism. The observed Trp blue shifts and K SV values (Table 2) indicate that for all of the (KW) n peptides tested, the Trp residues partitioned into the lipid bilayer of PE:PG (7:3, w/w) vesicles. The smallest peptide of the series, (KW)2, induced no calcein leakage or aggregation (Fig. 6), which likely reflects this peptide’s inability to permeate the bacterial membrane. (KW)3 exhibited a greater ability to bind LPS and induce membrane permeabilization, calcein leakage and aggregation than (KW)2, but its effects were still considerably weaker than those of (KW)4 and (KW)5. Furthermore, (KW)2 and (KW)3 lacked definite secondary structure on lipid membranes, as evidenced by their CD spectra. Briefly, the CD spectra showed that at a P/L ratio of 1:50, (KW)4 and (KW)5 formed stable structures within the negatively charged lipid membrane, suggesting interfacial association of the peptides (Fig. 7). Clearly, peptide length, hydrophobicity and charged/hydrophobic surface ratio correlate with the degree of membrane disruption/permeabilization. The exception is (KW)5, which showed less antibacterial activity than (KW)4, though its chain length is longer. This may reflect the weak aggregation of (KW)5 in aqueous solution, which would reduce its electrostatic potential and therefore its antibacterial activity. Our Trp blue shift assays and quenching experiments showed that its aggregation state led (KW)5 to bind more weakly to membranes than did (KW)4, which would also reduce its ability to permeabilize the membrane. Consistent with this interpretation, the degree of membrane disruption is reportedly dependent on the concentration of peptide adsorbed onto the negatively charged lipid membrane (Ringstad et al. 2007).
Many Trp-rich AMPs are not cell type-selective (Subbalakshmi et al. 1996; Ahmad et al. 1995; Schibli et al. 2002). In the present study, only (KW)5 caused significant leakage from PC:CH liposomes, and only (KW)5 exhibited a large blue shift, which indicates that only (KW)5 entered the hydrophobic core of the mammalian membrane. Our results also show that (KW) n peptides bind more tightly to bacterial membrane-mimicking liposomes than to those mimicking a mammalian membrane. This is because bacterial membranes consist of almost 25 % anionic lipids, whereas mammalian membranes contain large amounts of PC lipids, with negligible amounts of anionic lipids (Verkleij et al. 1973). Anionic lipids promote the initial electrostatic interaction between positively charged AMPs and the negatively charged membrane. Because the mammalian membrane is deficient in anionic lipids, AMPs cannot undergo a proper initial interaction. Infact, the positive charge of PC lipids may actually repulse the basic residues of AMPs. These results are consistent with an earlier study (Liu et al. 2007), which confirmed that (RW) n peptides strongly interact with PG-containing bilayers, but possess only moderate affinity toward mammalian model membranes. However, it is the hydrophobic interaction between AMPs and the lipid bilayer core that is essential for membrane disruption. Consequently, it is the more hydrophobic AMPs that lack cell type selectivity and cause more damage to membranes (Tossi et al. 2000; Makovitzki et al. 2006). Similarly, highly aggregated peptides having high hydrophobicity reportedly induce fatal damage to mammalian membranes (Biggs et al. 2007). The hemolysis and evoked calcein release seen with (KW)5 are consistent with that finding.
In addition to causing membrane damage, (KW)4 also elicited bacterial agglutination. Liposome agglutination is dependent on the number of electrostatic/hydrophobic interactions with the peptides, which in turn depends on the number of Lys/Trp residues (Stromstedt et al. 2009). Our data show that several Lys and Trp residues are required for permeabilization of the bacterial membrane and agglutination of model liposomes. Acrylamide quenching studies corroborate those results, as longer peptides permeated deeper into the lipid bilayer core than shorter ones (Table 2). The formation of macroscopic aggregates involves at least two steps: peptide-membrane association with concomitant charge neutralization followed by vesicle association (Cummings and Vanderlick 2007; Fujii et al. 1993). In addition to altering surface charge density upon adsorption to the membrane, peptides dehydrate lipid head groups and induce aggregation of phospholipid vesicles. Reducing charge density also reduces peptide–peptide long-distance repulsion, which facilitates the formation of hydrophobic interactions between peptides associated with adjacent bilayers (De Meulenaer et al. 1997). Other studies found that more highly charged residues, and neutralization of those charges may be required to induce fusion between giant liposomes (Nomura et al. 2004). It has also been reported that membrane destabilization is essential for fusion (Ryan et al. 2005 and Domingues et al. 2009), and that peptides and proteins forming hydrophobic or hydrophilic cationic clusters show membrane-fusion activity (Cummings and Vanderlick 2007; Niidome et al. 2000; Fujii et al. 1993; Abu-Ghazaleh et al. 1992). In the present study, (KW)4 and (KW)5 damaged membranes and caused vesicles aggregation/fusion (Fig. 6). Because (KW) n peptides contain only Lys and Trp residues, they can engage in both electrostatic and hydrophobic interactions with anionic lipid membranes and induce membrane destabilization and agglutination when dispersed on negatively charged vesicles.
The ability to cause vesicle leakage and aggregation was first identified with seminal ribonuclease (Mancheno et al. 1994) and various other synthetic peptides (Stromstedt et al. 2009), including K7, R7, K7W7 and R7W7. In the present study, (KW)4 destroyed the E. coli membrane (Supplementary Fig. 1) and caused agglutination of the bacterial cells (Supplementary Fig. 2). In addition to cell–cell aggregation, an extracellular matrix-like material was observed within the cell agglomeration. Proteins or peptides do not induce spontaneous aggregation of bacterial cells (Jung et al. 2009). Instead, agglutination results from peptide–charge and peptide–lipid interactions, which most likely involves hydrophilic/hydrophobic interactions with bacterial membranes (Gorr et al. 2008). Our finding that LPS-deficient Gram-positive bacteria, as well as liposomes, showed agglutination upon peptide treatment suggests that LPS does not play a role in bacterial agglutination.
Other studies have shown that RNase 3 triggers vesicle aggregation followed by membrane disruption, whereas RNase 7 induces dye leakage before aggregation (Torrent et al. 2009). Our SYTOX Green assays showed that (KW)4 and (KW)5 caused membrane destabilization at their MIC. In addition, the membrane depolarization assays showed that the peptides induced outer membrane permeabilization at concentrations lower than the MIC, and the time-frame of the effects of (KW)4 and (KW)5 on membrane permeability was consistent with the peptides’ bactericidal activities. However, (KW)4 induced bacterial agglutination at significantly higher concentrations than were needed to cause membrane permeabilization and bactericidal activity. This suggests that cell aggregation occurs after bacterial killing, and the initial step in the mechanism involves membrane disruption. Consistent with that idea, it is widely believed that membrane disruption is the primary mechanism by which AMPs exert their bactericidal effects (Hancock 2001; Brogden 2005; Tossi et al. 2000; Shai 2002). Induction of agglutination would help to clump bacterial cells together, which would increase their susceptibility to phagocytosis (Gorr et al. 2008). The ability of (KW)4 to bind to the bacterial membrane and induce agglutination makes it a potential candidate for future optimization.
In conclusion, the steps of the proposed mechanism by which (KW) n peptides exert their bactericidal effects are as follows (Fig. 8): (1) The peptide binds to membrane phospholipid polar heads through electrostatic interactions. (2) The hydrophobic residues promote peptide–lipid interactions, the clustering of cell/liposomes–peptide complexes and destabilization of the lipid bilayer. (3) Cell/liposome aggregation. Membranolytic peptides with agglutinating properties may eventually represent as a new class of short antibacterial peptides useful for killing bacteria resistant to other types of antibiotics.
Schematic representation of the proposed mechanism of action of (KW) n peptides in bacterial cells. (1) The peptide molecules associate with cell membranes or lipid bilayers, (2) the membrane/bilayer is disrupted, releasing the internal contents of the cell/vesicles; and (3) although higher peptide-lipid ratios necessary for cells/vesicles aggregate
Abbreviations
- CD:
-
Circular dichroism
- CH:
-
Cholesterol
- CFU:
-
Colony forming units
- DiSC3-5:
-
3,3′-Diethylthiodicarbocyanine iodide
- PC:
-
l-α-Phosphatidylcholine
- PE:
-
l-α-Phosphatidylethanolamine
- PG:
-
l-α-Phosphatidylglycerol
- Fmoc:
-
Fluoren-9-ylmethoxycarbonyl
- hRBCs:
-
Human red blood cells
- MIC:
-
Minimal inhibitory concentration
- PMB:
-
Polymyxin B
- SUVs:
-
Small unilamellar vesicles
- TAMRA:
-
5-Carboxytetramethylrhodamine
References
Abu-Ghazaleh RI, Gleich GJ, Prendergast FG (1992) Interaction of eosinophil granule major basic protein with synthetic lipid bilayers: a mechanism for toxicity. J Membr Biol 128:153–164
Ahmad I, Perkins WR, Lupan DM, Selsted ME, Janoff AS (1995) Liposomal entrapment of the neutrophil-derived peptide indolicidin endows it with in vivo antifungal activity. Biochim Biophys Acta 1237:109–114
Alanis AJ (2005) Resistance to antibiotics: are we in the post-antibiotic era? Arch Med Res 36:697–705
Andra J, Monreal D, Martinez de Tejada G, Olak C, Brezesinski G, Gomez SS, Goldmann T, Bartels R, Brandenburg K, Moriyon I (2007) Rationale for the design of shortened derivatives of the NK-lysine derived antimicrobial peptide NK-2 with improved activity against Gram-negative pathogens. J Biol Chem 282:14719–14728
Bhattachariya S, Ramamoorthy A (2009) Multifunctional host defense peptides: functional and mechanistic insights from NMR structures of potent antimicrobial peptides. FEBS J 276:6465–6473
Biggs JS, Rosenfeld Y, Shai Y, Olivera BM (2007) Conolysin-Mt: a Conus peptide that disrupts cellular membranes. Biochemistry 46:12586–12593
Brogden KA (2005) Antimicrobial peptides: pore formers or metabolic inhibitors in bacteria? Nat Rev Microbiol 3:238–250
Brogden NK, Brogden KA (2011) Will new generations of modified antimicrobial peptides improve their potential as pharmaceuticals? Int J Antimicrob Agents 38:217–225
Brown KL, Hancock RE (2006) Cationic host defense (antimicrobial) peptides. Curr Opin Immunol 18:24–30
Burkert U, Allinger NL (1982) Molecular mechanics. ACS Monograph N. 177. ACS, Washington, DC, p 339
Burstein EA, Vendenkina NS, Ivkova MN (1973) Fluorescence and the location of tryptophan residues in protein molecules. Photochem Photobiol 18:263–279
Cummings JE, Vanderlick TK (2007) Aggregation and hemifusion of anionic vesicles induced by the antimicrobial peptide cryptdin-4. Biochim Biophys Acta 1768:1796–1804
Dathe M, Nikolenko H, Klose J, Bienert M (2004) Cyclization increases the antimicrobial activity and selectivity of arginine- and tryptophan-containing hexapeptides. Biochemistry 43:9140–9150
De Meulenaer B, Van der Meeren P, De Cuyper M, Vanderdeelen J, Baert L (1997) Electrophoretic and dynamic light scattering study of the interaction of cytochrome, C with dimyristoylphosphatidylglycerol, dimyristoylphosphatidylcholine, and intramembranously mixed liposomes. J Colloid Interface Sci 189:254–258
DeLano WL (2002) The PyMol molecular graphics system. Delano Scientific, San Carlos
Deslouches B, Phadke SM, Lazarevic V, Cascio M, Islam K, Montelaro RC, Mietzner TA (2005) De novo generation of cationic antimicrobial peptides: influence of length and tryptophan substitution on antimicrobial activity. Antimicrob Agents Chemother 49:316–322
Domingues MM, Castanho MA, Santos NC (2009) rBPI21 promotes lipopolysaccharide aggregation and exerts its antimicrobial effects by (hemi) fusion of PG-containing membranes. PLoS ONE 4:e8385
Epand RF, Lehrer RI, Waring A, Wang W, Maget-Dana R, Lelievre D, Epand RM (2003) Direct comparison of membrane interactions of model peptides composed of only Leu and Lys residues. Biopolymers 71:2–16
Farnaud S, Spiller C, Moriarty LC, Patel A, Gant V, Odell EW, Evans RW (2004) Interactions of lactoferricin-derived peptides with LPS and antimicrobial activity. FEMS Microbiol Lett 233:193–199
Feder R, Dagan A, Mor A (2000) Structure–activity relationship study of antimicrobial dermaseptin S4 showing the consequences of peptide oligomerization on selective cytotoxicity. J Biol Chem 275:4230–4238
Fernandez-Lopez S, Kim HS, Choi EC, Delgado M, Granja JR, Khasanov A, Kraehenbuehl K, Long G, Weinberger DA, Wilcoxen KM, Ghadiri MR (2001) Antibacterial agents based on the cyclic d, l-α-peptide architecture. Nature 412:452–455
Fjell CD, Hiss JA, Hancock REW, Schneider G (2011) Designing antimicrobial peptides: form follows function. Nat Rev Drug Discov 11:37–51
Fujii G, Selsted ME, Eisenberg D (1993) Defensins promote fusion and lysis of negatively charged membranes. Protein Sci 2:1301–1312
Ghosh JK, Shaool D, Guillaud P, Ciceron L, Mazier D, Kustanovich I, Shai Y, Mor A (1997) Selective cytotoxicity of dermaseptin S3 toward intraerythrocytic Plasmodium falciparum and the underlying molecular basis. J Biol Chem 272:31609–31616
Giuliani A, Rinaldi AC (2011) Beyond natural antimicrobial peptides: multimeric peptides and other peptidomimetic approaches. Cell Mol Life Sci 68:2255–2266
Gopal R, Park SC, Ha KJ, Cho SJ, Kim SW, Song PI, Nah JW, Park Y, Hahm KS (2009) Effect of leucine and lysine substitution on the antimicrobial activity and evaluation of the mechanism of the HPA3NT3 analog peptide. J Pept Sci 15:589–594
Gopal R, Kim YJ, Seo CH, Hahm KS, Park Y (2011) Reversed sequence enhances antimicrobial activity of a synthetic peptide. J Pept Sci 17:329–334
Gorr SU, Sotsky JB, Shelar AP, Demuth DR (2008) Design of bacteria-agglutinating peptides derived from parotid secretory protein, a member of the bactericidal/permeability increasing-like protein family. Peptides 29:2118–2127
Hancock RE (2001) Cationic peptides: effectors in innate immunity and novel antimicrobials. Lancet Infect Dis 1:156–164
Javadpour MM, Barkley MD (1997) Self-assembly of designed antimicrobial peptides in solution and micelles. Biochemistry 36:9540–9549
Javadpour MM, Juban MM, Lo WC, Bishop SM, Alberty JB, Cowell SM, Becker CL, McLaughlin ML (1996) De novo antimicrobial peptides with low mammalian cell toxicity. J Med Chem 39:3107–3113
Juffer AH, Shepherd CM, Vogel HJ (2001) Protein-membrane electrostatic interactions: applications of the lekner summation technique. J Chem Phys 114:1892–1905
Jung S, Dingley AJ, Augustin R, Erxleben FA, Stanisak M, Gelhaus C, Gutsmann T, Hammer MU, Podschun R, Bonvin AM, Leippe M, Bosch TC, Grotzinger J (2009) Hydramacin-1, structure and antibacterial activity of a protein from the basal metazoan hydra. J Biol Chem 284:1896–1905
Kim JY, Park SC, Yoon MY, Hahm KS, Park Y (2011) C-terminal amidation of PMAP-23: translocation to the inner membrane of Gram-negative bacteria. Amino Acids 40:183–195
Kiyota T, Lee S, Sugihara G (1996) Design and synthesis of amphiphilic α-helical model peptides with systematically varied hydrophobic-hydrophilic balance and their interaction with lipid- and bio-membranes. Biochemistry 35:13196–13204
Ladokhin AS, Selsted ME, White SH (1997) Bilayer interactions of indolicidin, a small antimicrobial peptide rich in tryptophan, proline, and basic amino acids. Biophys J 72:794–805
Liu Z, Brady A, Young A, Rasimick B, Chen K, Zhou C, Kallenbach NR (2007) Length effects in antimicrobial peptides of the (RW) n series. Antimicrob Agents Chemother 51:597–603
Lugtenberg B, Van Alphen L (1983) Molecular architecture and functioning of the outer membrane of Escherichia coli and other Gram-negative bacteria. Biochim Biophys Acta 737:51–115
Makovitzki A, Avrahami D, Shai Y (2006) Ultrashort antibacterial and antifungal lipopeptides. Proc Natl Acad Sci USA 103:15997–16002
Mancheno JM, Gasset M, Onederra M, Gavilanes JG, Dalessio G (1994) Bovine seminal ribonuclease destabilizes negatively charged membranes. Biochem Biophys Res Commun 199:119–124
Mao D, Wallace BA (1984) Differential light scattering and adsorption flattening optical effects are minimal in the circular dichroism spectra of small unilamellar vesicles. Biochemistry 23:2667–2673
Matsuzaki K, Sugishita K, Miyajima K (1999) Interactions of an antimicrobial peptide, magainin 2, with lipopolysaccharide-containing liposomes as a model for outer membranes of Gram-negative bacteria. FEBS Lett 449:221–224
Mayer LD, Hope MJ, Cullis PR (1986) Vesicles of variable sizes produced by a rapid extrusion procedure. Biochim Biophys Acta 858:161–168
Moore RA, Bates NC, Hancock RE (1986) Interaction of polycationic antibiotics with Pseudomonas aeruginosa lipopolysaccharide and lipid A studied by using dansyl-polymyxin. Antimicrob Agents Chemother 29:496–500
Niidome T, Tsuiki M, Tokunaga Y, Hatakeyama T, Aoyagi H (2000) Antibacterial activity of Arg/Pro-rich bactenecin 5 model peptides and their interaction with phospholipid membranes. Bull Chem Soc Jpn 73:1397–1402
Niidome T, Matsuyama N, Kunihara M, Hatakeyama T, Aoyagi H (2005) Effect of chain length of cationic model peptides on antibacterial activity. Bull Chem Soc Jpn 78:473–476
Nomura F, Inaba T, Ishikawa S, Nagata M, Takahashi S, Hotani H, Takiguchi K (2004) Microscopic observations reveal that fusogenic peptides induce liposome shrinkage prior to membrane fusion. Proc Natl Acad Sci USA 101:3420–3425
Oren Z, Lerman JC, Gudmundsson GH, Agerberth B, Shai Y (1999) Structure and organization of the human antimicrobial peptide LL-37 in phospholipid membranes: relevance to the molecular basis for its non-cell-selective activity. Biochem J 341:501–513
Papo N, Oren Z, Pag U, Sahl HG, Shai Y (2002) The consequence of sequence alternation of an amphipathic α-helical antimicrobial peptide and its diastereomers. J Biol Chem 277:33913–33921
Park Y, Lee DG, Jang SH, Woo ER, Jeong HG, Choi CH, Hahm KS (2003) A Leu Lys-rich antimicrobial peptide: activity and mechanism. Biochim Biophys Acta 1645:172–182
Powers JP, Hancock RE (2003) The relationship between peptide structure and antibacterial activity. Peptides 24:1681–1691
Ringstad L, Nordahl AE, Schmidtchen A, Malmsten M (2007) Composition effect on peptide interaction with lipids and bacteria: variants of C3a peptide CNY21. Biophys J 92:87–98
Rosenfeld Y, Lev N, Shai Y (2010) Effect of the hydrophobicity to net positive charge ratio on antibacterial and anti-endotoxin activities of structurally similar antimicrobial peptides. Biochemistry 49:853–861
Ryan MA, Qi X, Serrano AG, Ikegami M, Gil JP, Johansson J, Weaver TE (2005) Mapping and analysis of the lytic and fusogenic domains of surfactant protein B. Biochemistry 44:861–872
Sal-Man N, Oren Z, Shai Y (2002) Preassembly of membrane-active peptides is an important factor in their selectivity toward target cells. Biochemistry 41:11921–11930
Schibli DJ, Epand RF, Vogel HJ, Epand RM (2002) Tryptophan-rich antimicrobial peptides: comparative properties and membrane interactions. Biochem Cell Biol 80:667–677
Shai Y (2002) Mode of action of membrane active antimicrobial peptides. Biopolymers 66:236–248
Shepherd CM, Schaus KA, Vogel HJ, Juffer AH (2001) Molecular dynamics study of peptide-bilayer adsorption. Biophys J 80:579–596
Song YM, Park Y, Lim SS, Yang ST, Woo ER, Park IS, Lee JS, Kim JI, Hahm KS, Kim Y, Shin SY (2005) Cell selectivity and mechanism of action of antimicrobial model peptides containing peptoid residues. Biochemistry 44:12094–12106
Stewart JC (1980) Colorimetric determination of phospholipids with ammonium ferrothiocyanate. Anal Biochem 104:10–14
Strom MB, Haug BE, Rekdal O, Skar ML, Stensen W, Svendsen JS (2002) Important structural features of 15-residue lactoferricin derivatives and methods for improvement of antibacterial activity. Biochem Cell Biol 80:65–74
Strom MB, Haug BE, Skar ML, Stensen W, Stiberg T, Svendsen JS (2003) The pharmacophore of short cationic antibacterial peptides. J Med Chem 46:1567–1570
Stromstedt AA, Pasupuleti M, Schmidtchen A, Malmsten M (2009) Oligotryptophan-tagged antimicrobial peptides and the role of the cationic sequence. Biochim Biophys Acta 1788:1916–1923
Subbalakshmi C, Krishnakumari V, Nagaraj R, Sitaram N (1996) Requirements for antibacterial and hemolytic activities in the bovine neutrophil derived 13-residue peptide indolicidin. FEBS Lett 395:48–52
Subbalakshmi C, Bikshapathy E, Sitaram N, Nagaraj R (2000) Antibacterial and hemolytic activities of single tryptophan analogs of indolicidin. Biochem Biophys Res Commun 274:714–716
Tenover FC (2006) Mechanisms of antimicrobial resistance in bacteria. Am J Med Sci 119:S3–S10
Torrent M, Sanchez D, Buzon V, Nogues MV, Cladera J, Boix E (2009) Comparison of the membrane interaction of two antimicrobial RNases: RNase 3/ECP and RNase 7. Biochim Biophys Acta 1788:1116–1125
Tossi A, Sandri L, Giangaspero A (2000) Amphipathic, α-helical antimicrobial peptides. Biopolymers 55:4–30
Verkleij AJ, Zwaal RF, Roelofsen B, Comfurius P, Kastelijin D, Van Deenen LL (1973) The asymmetric distribution of phospholipids in the human red cell membrane. A combined study using phospholipases and freeze-etch electron microscopy. Biochim Biophys Acta 323:178–193
Wimley WC, White SH (1996) Experimentally determined hydrophobicity scale for proteins at membrane interfaces. Nat Struct Biol 3:842–848
Wimley WC, Creamer TP, White SH (1996) Solvation energies of amino acid side chains and backbone in a family of host–guest pentapeptides. Biochemistry 35:5109–5124
Woody RW (1994) Contributions of tryptophan side chains to the far-ultraviolet circular dichroism of proteins. Eur Biophys J 23:253–262
Yang ST, Shin SY, Lee CW, Kim YC, Hahm KS, Kim JI (2003) Selective cytotoxicity following Arg-to-Lys substitution in tritrpticin adopting a unique amphipathic turn structure. FEBS Lett 540:229–233
Zilberstein D, Schuldiner S, Padan E (1979) Proton electrochemical gradient in Escherichia coli cells and its relation to active transport of lactose. Biochemistry 18:669–673
Acknowledgments
This work was supported by the National Research Foundation of Korea (NRF) grant funded by the Korea government (MEST) (No. 2011-0017532).
Open Access
This article is distributed under the terms of the Creative Commons Attribution License which permits any use, distribution, and reproduction in any medium, provided the original author(s) and the source are credited.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 2.0 International License (https://creativecommons.org/licenses/by/2.0), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
About this article
Cite this article
Gopal, R., Seo, C.H., Song, P.I. et al. Effect of repetitive lysine–tryptophan motifs on the bactericidal activity of antimicrobial peptides. Amino Acids 44, 645–660 (2013). https://doi.org/10.1007/s00726-012-1388-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00726-012-1388-6