Abstract
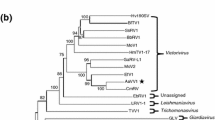
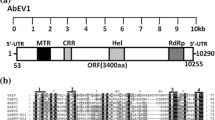
The phytopathogenic fungus Alternaria spp. contains a variety of double-stranded RNA (dsRNA) elements of different sizes. Detailed analysis of next-generation sequencing data obtained using dsRNA purified from Alternaria arborescens, from which we had previously found Alternaria arborescens victorivirus 1, revealed the presence of another mycoviral-like dsRNA of approximately 2.5 kbp in length. When using the fungal mitochondrial genetic code, this dsRNA has a single open reading frame that potentially encodes an RNA-dependent RNA polymerase (RdRp) with significant to sequence similarity to those of viruses of the genus Mitovirus. Moreover, both the 5’- and 3’-untranslated regions have the potential to fold into stable stem-loop structures, which is characteristic of mitoviruses. Pairwise comparisons and phylogenetic analysis of the deduced amino acid sequences of RdRp indicated that the virus we identified in A. arborescens is a distinct member of the genus Mitovirus in the family Narnaviridae, designated as “Alternaria arborescens mitovirus 1” (AaMV1).

Similar content being viewed by others
References
Aoki N, Moriyama H, Kodama M, Arie T, Teraoka T, Fukuhara T (2009) A novel mycovirus associated with four double-stranded RNAs affects host fungal growth in Alternaria alternata. Virus Res 140:179–187
Botella L, Tuomivirta TT, Vervuurt S, Diez JJ, Hantula J (2012) Occurrence of two different species of mitoviruses in the European race of Gremmeniella abietina var. abietina, both hosted by the genetically unique Spanish population. Fungal Biol 116:872–882
Deng F, Boland GJ (2006) Attenuation of virulence in Sclerotinia homoeocarpa during storage is associated with latent infection by Ophiostoma mitovirus 3a. Eur J Plant Pathol 114:127–137
Fuke K, Takeshita K, Aoki N, Fukuhara T, Egusa M, Kodama M, Moriyama H (2011) The presence of double-stranded RNA in Alternaria alternata Japanese pear pathotype is associated with morphological changes. J Gen Plant Pathol 77:248–252
Ghabrial SA, Castón JR, Jiang D, Nibert ML, Suzuki N (2015) 50-plus years of fungal viruses. Virology 479–480:356–368
Hillman BI, Cai G (2013) The family Narnaviridae: simplest of RNA viruses. Adv Virus Res 86:149–176
Khalifa ME, Varsani A, Ganley AR, Pearson MN (2015) Comparison of Illumina de novo assembled and Sanger sequenced viral genomes: A case study for RNA viruses recovered from the plant pathogenic fungus Sclerotinia sclerotiorum. Virus Res. doi:10.1016/j.virusres.2015.11.001
King AMQ, Lefkowitz E, Adams MJ, Carstens B (2012) Virus taxonomy: ninth report of the International Committee on Taxonomy of Viruses. Elsevier, Amsterdam
Komatsu K, Fukuhara T, Arie T, Teraoka T, Omatsu T, Mizutani T, Kodama M, Moriyama H (2016) Genome sequence of a novel victorivirus identified in the phytopathogenic fungus Alternaria arborescens. Arch Virol. doi:10.1007/s00705-016-2796-9
Koonin EV, Dolja VV, Krupovic M (2015) Origins and evolution of viruses of eukaryotes: the ultimate modularity. Virology 479–480:2–25
Lin Y, Zhang H, Zhao C, Liu S, Guo L (2015) The complete genome sequence of a novel mycoviurs from Alternaria longipes strain HN28. Arch Virol 160:577–580
Okada R, Kiyota E, Moriyama H, Fukuhara T, Natsuaki T (2015) A simple and rapid method for viral dsRNA purification from plant and fungal tissue. J Gen Plant Pathol 81:103–107
Reuter JS, Mathews DH (2010) RNAstructure: software for RNA secondary structure prediction and analysis. BMC Bioinfom 11:129
Shang HH, Zhong J, Zhang RJ, Chen CY, Gao BD, Zhu HJ (2015) Genome sequence of a novel endornavirus from the phytopathogenic fungus Alternaria brassicicola. Arch Virol 160:1827–1830
Tamura K, Stecher G, Peterson D, Filipski A, Kumar S (2013) MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol 30:2725–2729
Xie J, Ghabrial SA (2012) Molecular characterizations of two mitoviruses co-infecting a hypovirulent isolate of the plant pathogenic fungus Sclerotinia sclerotiorum. Virology 428:77–85
Acknowledgments
We thank T. Peever (Washington State University, USA) for providing the A. arborescens strain.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Funding
This study was funded by a Grant-in-Aid for Scientific Research (C) (15K07838) from the Japan Society for the Promotion of Science.
Conflict of interest
The authors declare no conflict of interest.
Ethical approval
This article does not contain any studies with animals and human participants performed by any of the authors.
Electronic supplementary material
Below is the link to the electronic supplementary material.
705_2016_2953_MOESM1_ESM.pptx
Fig. S1 Multiple alignment of the deduced amino acid (aa) sequence of the RdRp encoded by AaMV1 and selected mitoviruses. The positions of the six conserved motifs in RdRps (I-VI) of dsRNA viruses are indicated by lines above the alignment. Asterisks and dots below the alignment indicate identical and conserved aa residues, respectively. Numbers at the beginning of the alignment represent the aa position from the start of the predicted gene product, and those in square brackets represent the number of aa residues between the motifs (PPTX 1581 kb)
705_2016_2953_MOESM2_ESM.pptx
Fig. S2 Predicted secondary structures of the 5’- (a) and 3’- (b) UTRs of AbMV1 using the RNAstructure program. ∆G values, which represent the free energy, are indicated below each structure. Nucleotide numbers of the AbMV1 genome are also indicated (PPTX 54 kb)
Rights and permissions
About this article
Cite this article
Komatsu, K., Katayama, Y., Omatsu, T. et al. Genome sequence of a novel mitovirus identified in the phytopathogenic fungus Alternaria arborescens . Arch Virol 161, 2627–2631 (2016). https://doi.org/10.1007/s00705-016-2953-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00705-016-2953-1