Abstract
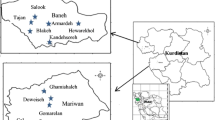
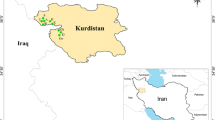
Alfalfa (Medicago sativa subsp. sativa) was introduced to Lithuania as a fodder crop and now grows as a weed in natural ecosystems adjacent to its wild relative, sickle medic (M. sativa subsp. falcata). The hybrid form of these taxa, variegated alfalfa (M. sativa subsp. varia), is considered an invasive taxon due to its intensive spread. Here, we use inter-simple sequence repeat (ISSR) markers and haplotype analyses of three regions of chloroplast DNA (psbA-trnH, trnS-trn2GS and trnL-trnF) and the nuclear ribosomal DNA internal transcribed spacer region to study the spontaneous hybridization of alfalfa and sickle medic in natural subpopulations. Plants of the three taxa were collected from 19 subpopulations located at seven sites. ISSR analysis revealed similar values of genetic diversity (expected heterozygosity, band richness, polymorphic band proportion) in subpopulations of the three studied subspecies. Our study revealed a lack of genetic differentiation among these taxa and high differentiation among subpopulations (Φ ST = 0.389, P = 0.001). Analysis of the chloroplast DNA regions provides direct evidence for the hybrid origin of variegated alfalfa. Taxon-characteristic haplotypes were identified for subsp. sativa and subsp. falcata in local subpopulations, and two haplotypes specific to the parental taxa were detected in subpopulations of M. sativa subsp. varia. Our study demonstrates the extensive genetic impact of alfalfa on adjacent native subpopulations of sickle medic.

Similar content being viewed by others
References
Abbott RJ (1992) Plant invasions, interspecific hybridization and the evolution of new plant taxa. Trends Ecol Evol 7:401–405. doi:10.1016/0169-5347(92)90020-C
Blair MW, Panaud O, McCouch SR (1999) Inter-simple sequence repeat (ISSR) amplification for analysis of microsatellite motif frequency and fingerprinting in rice (Oryza sativa L.). Theor Appl Genet 98:780–792. doi:10.1007/s001220051135
Bleeker W, Schmitz U, Ristow M (2007) Interspecific hybridisation between alien and native plant species in Germany and its consequences for native biodiversity. Biol Conservation 137:248–253. doi:10.1016/j.biocon.2007.02.004
Bonin A, Bellemain E, Bronken Eidesen P, Pompanon F, Brochmann C, Taberlet P (2004) How to track and assess genotyping errors in population genetics studies. Molec Ecol 13:3261–3273. doi:10.1111/j.1365-294X.2004.02346.x
Celka Z, Szczecińska M, Sawicki J, Shevera MV (2012) Molecular studies did not support the distinctiveness of Malva alcea and M. excisa (Malvaceae) in Central and Eastern Europe. Biologia (Bratislava) 67:1088–1098. doi:10.2478/s11756-012-0107-9
Coart E, Van Glabeke S, Petit RJ, Van Bockstaele E, Roldán-Ruiz I (2005) Range wide versus local patterns of genetic diversity in hornbeam (Carpinus betulus L.). Conservation Genet 6:259–273. doi:10.1007/s10592-004-7833-7
Crochemore ML, Huyghe C, Écalle C, Julier B (1998) Structuration of alfalfa genetic diversity using agronomic and morphological characteristics. Relationship with RAPD markers. Agronomie 18:79–94
Earl DA, vonHoldt BM (2012) STRUCTURE HARVESTER: a website and program for visualizing STRUCTURE output and implementing the Evanno method. Conservation Genet Resour 4:359–361. doi:10.1007/s12686-011-9548-7
Ellstrand NC, Schierenbeck K (2000) Hybridization as a stimulus for the evolution of invasiveness in plants? Proc Natl Acad Sci USA 97:7043–7050. doi:10.1073/pnas.97.13.7043
Ellstrand NC, Prentice HC, Hancock JF (1999) Gene flow and introgression from domesticated plants into their wild relatives. Annual Rev Ecol Syst 30:539–563. doi:10.1146/annurev.ecolsys.30.1.539
Excoffier L, Smouse PE, Quattro JM (1992) Analysis of molecular variance inferred from metric distances among DNA haplotypes: application to human mitochondria DNA restriction sites. Genetics 131:479–491
Fajardo CG, de Almeida Vieira F, Molina WF (2014) Interspecific genetic analysis of orchids in Brazil using molecular markers. Pl Syst Evol 300:1825–1832. doi:10.1007/s00606-014-1009-9
Falush D, Stephens M, Pritchard JK (2007) Inference of population structure using multilocus genotype data: dominant markers and null alleles. Molec Ecol Notes 7:574–578. doi:10.1111/j.1471-8286.2007.01758.x
Ge XJ, Yua Y, Zhao NX, Chen HS, Qi WQ (2003) Genetic variation in the endangered Inner Mongolia endemic shrub Tetraena mongolica Maxim. (Zygophyllaceae). Biol Conservation 111:427–434. doi:10.1016/S0006-3207(02)00312-9
Geleta M, Bryngelsson T (2012) Population genetic analysis of Lobelia rhynchopetalum Hemsl. (Campanulaceae) using DNA sequences from ITS and eight chloroplast DNA regions. Sci World J 2012:276451. doi:10.1100/2012/276451
Havananda T, Brummer EC, Doyle JJ (2011) Complex patterns of autopolyploid evolution in alfalfa and allies (Medicago sativa; Leguminosae). Amer J Bot 98:1633–1646. doi:10.3732/ajb.1000318
Hollingsworth PM, Graham SW, Little DP (2011) Choosing and using a plant DNA barcode. PLoS One 6:e19254. doi:10.1371/journal.pone.0019254
Jenczewski E, Prosperi JM, Ronfort J (1999) Differentiation between natural and cultivated populations of Medicago sativa (Leguminosae) from Spain: analysis with random amplified polymorphic DNA (RAPD) markers and comparison to allozymes. Molec Ecol 8:1317–1330. doi:10.1046/j.1365-294X.1999.00697.x
Kaljund K, Leht M (2010) Medicago falcata L. in Estonia: chromosomal and morphological variability, distribution and vulnerability of taxa. Acta Biol Univ Daugavpils S2:107–119
Kaljund K, Leht M (2013) Extensive introgressive hybridization between cultivated lucerne and the native sickle medic (Medicago sativa ssp. falcata) in Estonia. Ann Bot Fenn 50:23–31
Kupcinskiene E, Zybartaite L, Janulioniene R, Zukauskiene J, Paulauskas A (2013) Molecular diversity of small balsam populations in relation to site characteristics. Centr Eur J Biol 8:1048–1061. doi:10.2478/s11535-013-0228-3
Lapiņa L, Grauda D, Rashal I (2011) Characterization of Latvian alfalfa Medicago sativa genetic resources. Acta Biol Univ Daugavpils 11:134–140
Lee DJ, Blake TK, Smith SE (1988) Biparental inheritance of chloroplast DNA and the existence of heteroplasmic cells in alfalfa. Theor Appl Genet 76:545–549. doi:10.1007/BF00260905
Levin DA, Francisco-Ortega J, Jansen RK (1996) Hybridization and the extinction of rare plant species. Conservation Biol 10:10–16. doi:10.1046/j.1523-1739.1996.10010010.x
Lynch M, Milligan BG (1994) Analysis of population genetic structure with RAPD markers. Molec Ecol 3:91–99. doi:10.1111/j.1365-294X.1994.tb00109.x
Mengoni A, Gori A, Bazzicalupo M (2000) Use of RAPD and microsatellite (SSR) variation to assess genetic relationships among populations of tetraploid alfalfa, Medicago sativa. Pl Breed 119:311–317. doi:10.1046/j.1439-0523.2000.00501.x
Michaud R, Lehman WF, Rumbaugh MD (1988) World distribution and historical development. In: Hanson AA, Barnes DK, Hill RR Jr (eds) Alfalfa and alfalfa improvement. ASA-CSSA-SSSA, Madison, WI, pp 25– 91
Morrell P, Rieseberg L (1998) Molecular tests of the proposed diploid hybrid origin of Gilia achilleifolia (Polemoniaceae). Amer J Bot 85:1439–1453
Muller MH, Prosperi JM, Santoni S, Ronfort J (2003) Inferences from mitochondrial DNA patterns on the domestication history of alfalfa (Medicago sativa). Molec Ecol 12:2187–2199. doi:10.1046/j.1365-294X.2003.01897.x
Naugžemys D, Žilinskaitė S, Skridaila A, Žvingila D (2014) Phylogenetic analysis of the polymorphic 4× species complex Lonicera caerulea (Caprifoliaceae) using RAPD markers and noncoding chloroplast DNA sequences. Biologia (Bratislava) 69:585–593. doi:10.2478/s11756-014-0345-0
Nybom H (2004) Comparison of different nuclear DNA markers for estimating intraspecific genetic diversity in plants. Molec Ecol 13:1143–1155. doi:10.1111/j.1365-294X.2004.02141.x
Oddou-Muratorio S, Petit RJ, Le Guerroué B, Guesnet D, Demesure B (2001) Pollen-versus seed-mediated gene flow in a scattered forest tree species. Evolution 55:1123–1135
Patamsytė J, Čėsnienė T, Naugžemys D, Kleizaitė V, Vaitkūnienė V, Rančelis V, Žvingila D (2011) Genetic diversity of warty cabbage (Bunias orientalis L.) revealed by RAPD and ISSR markers. Zemdirbyste 98:293–300
Peakall R, Smouse PE (2012) GenAlEx 6.5: genetic analysis in Excel. Population genetic software for teaching and research—an update. Bioinformatics 28:2537–2539. doi:10.1093/bioinformatics/bts460
Petit RJ, El Mousadik A, Pons O (1998) Identifying populations for conservation on the basis of genetic markers. Conservation Biol 12:844–855. doi:10.1111/j.1523-739.1998.96489.x
Petit RJ, Bodénès C, Ducousso A, Roussel G, Kremer A (2003) Hybridization as a mechanism of invasion in oaks. New Phytol 161:151–164. doi:10.1046/j.1469-8137.2003.00944.x
Piya S, Nepal MP, Butler JL, Larson GE, Neupane A (2014) Genetic diversity and population structure of sickleweed (Falcaria vulgaris; Apiaceae) in the upper Midwest USA. Biol Invasions 16:2115–2125. doi:10.1007/s10530-014-0651-z
Pleines T, Jakob SS, Blattner FR (2009) Application of non-coding DNA regions in intraspecific analyses. Pl Syst Evol 282:281–294. doi:10.1007/s00606-008-0036-9
Quiros CF, Bauchan GR (1988) The genus Medicago and the origin of the Medicago sativa complex. In: Hanson AA, Barnes DK, Hill RR (eds) Alfalfa and alfalfa improvement. American Society of Agronomy Inc., Madison, pp 93–124
Riday H, Brummer EC, Campbell TA, Luth D, Cazcarro PM (2003) Comparisons of genetic and morphological distance with heterosis between Medicago sativa subsp. sativa and subsp. falcata. Euphytica 131:37–45. doi:10.1023/A:1023050126901
Rosenberg NA (2004) DISTRUCT: a program for the graphical display of population structure. Molec Ecol Notes 4:137–138. doi:10.1046/j.1471-8286.2003.00566.x
Ross C, Auge H, Durka W (2008) Genetic relationships among three native North-American Mahonia species, invasive Mahonia populations from Europe, and commercial cultivars. Pl Syst Evol 275:219–229. doi:10.1007/s00606-008-0066-3
Schierenbeck KA, Ellstrand NC (2009) Hybridization and the evolution of invasiveness in plants and other organisms. Biol Invasions 11:1093–1105. doi:10.1007/s10530-008-9388-x
Schierenbeck KA, Symonds VV, Gallagher KG, Bell J (2005) Genetic variation and phylogeographic analyses of two species of Carpobrotus and their hybrids in California. Molec Ecol 14:539–547. doi:10.1111/j.1365-294X.2005.02417.x
Shaw J, Lickey EB, Beck JT, Farmer SB, Liu WS, Miller J, Siripun KC, Winder CT, Schilling EE, Small RL (2005) The tortoise and the hare II: relative utility of 21 noncoding chloroplast DNA sequences for phylogenetic analysis. Amer J Bot 92:142–166. doi:10.3732/ajb.92.1.142
Small E (2011) Alfalfa and relatives. Evolution and classification of Medicago. NRC Research Press, Ottawa
Small E, Brookes BS (1984) Taxonomic circumscription and identification in the Medicago sativa-falcata (Alfalfa) continuum. Econ Bot 38:83–96. doi:10.1007/BF02904419
Stebbins GL (1950) Variation and evolution in plants, 2nd edn. Columbia University Press, New York
Tamura K, Stecher G, Peterson D, Filipski A, Kumar S (2013) MEGA6: molecular evolutionary genetics analysis version 6.0. Molec Biol Evol 30:2725–2729. doi:10.1093/molbev/mst197
Taški-Ajduković K, Nagl N, Milić D, Katić S, Zorić M (2014) Genetic variation and relationship of alfalfa populations and their progenies based on RAPD markers. Centr Eur J Biol 9:768–776. doi:10.2478/s11535-014-0307-0
Tovar-Sánchez E, Rodríguez-Carmona F, Aguilar-Mendiola V, Mussali-Galante P, López-Caamal A, Valencia-Cuevas L (2012) Molecular evidence of hybridization in two native invasive species: Tithonia tubaeformis and T. rotundifolia (Asteraceae) in Mexico. Pl Syst Evol 298:1947–1959. doi:10.1007/s00606-012-0693-6
Vekemans X, Beauwens T, Lemaire M, Roldán-Ruiz I (2002) Data from amplified fragment length polymorphism (AFLP) markers show indication of size homoplasy and of a relationship between degree of homoplasy and fragment size. Molec Ecol 11:139–151. doi:10.1046/j.0962-1083.2001.01415.x
Vyšniauskienė R, Rančelienė V, Patamsytė J, Žvingila D (2013) High genetic differentiation among wild populations of alien Medicago sativa in Lithuania. Centr Eur J Biol 8:480–491. doi:10.2478/s11535-013-0159-4
Ward SM, Fleischmann CE, Turner MF, Sing SE (2009) Hybridization between invasive populations of Dalmatian toadflax (Linaria dalmatica) and yellow toadflax (Linaria vulgaris). Invasive Pl Sci Managm 2:369–378. doi:10.1614/IPSM-09-031.1
White TJ, Bruns T, Lee S, Taylor JW (1990) Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis MA, Gelfand DH, Sninsky JJ, White TJ (eds) PCR protocols: a guide to methods and applications. Academic Press, New York, pp 315–322
Yeh FC, Yang RC, Boyle TBJ, Ye ZH, Mao JX (1997) POPGENE, the user-friendly shareware for population genetic analysis. Molecular Biology and Biotechnology Centre, University of Alberta, Canada
Acknowledgments
This research was founded by a grant (No. LEK-07/2012) from the Research Council of Lithuania. We thank associate editor W. Durka and three anonymous referees for valuable comments. We also thank M. Staniūtė for assistance in this study.
Author information
Authors and Affiliations
Corresponding author
Additional information
Handling editor: Walter Durka.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Vyšniauskienė, R., Naugžemys, D., Patamsytė, J. et al. ISSR and chloroplast DNA analyses indicate frequent hybridization of alien Medicago sativa subsp. sativa and native M. sativa subsp. falcata . Plant Syst Evol 301, 2341–2350 (2015). https://doi.org/10.1007/s00606-015-1232-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00606-015-1232-z