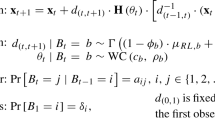Abstract
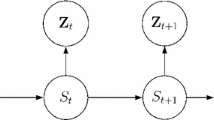
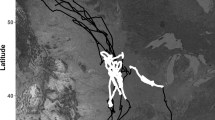
The modelling of animal movement is an important ecological and environmental issue. It is well-known that animals change their movement patterns over time, according to observable and unobservable factors. To trace the dynamics of behaviors, to identify factors influencing these dynamics and unobserved characteristics driving intra-subjects correlations, we introduce a time-dependent mixed effects projected normal regression model. A set of animal-specific parameters following a hidden Markov chain is introduced to deal with unobserved heterogeneity. For the maximum likelihood estimation of the model parameters, we outline an expectation–maximization algorithm. A large-scale simulation study provides evidence on model behavior. The data analysis approach based on the proposed model is finally illustrated by an application to a dataset, which derives from a population of Talitrus saltator from the beach of Castiglione della Pescaia (Italy).






Similar content being viewed by others
References
Bacci S, Pandolfi A, Pennoni F (2014) A comparison of some criteria for states selection in the latent Markov model for longitudinal data. Adv Data Anal Classif 8:125–145
Bartolucci F, Farcomeni A (2009) A multivariate extension of the dynamic logit model for longitudinal data based on a latent Markov heterogeneity structure. J Am Stat Assoc 104:816–831
Bartolucci F, Farcomeni A, Pennoni F (2013) Latent Markov model for longitudinal data. CRC Press, Boca Raton
Breed GA, Jonsen ID, Myers RA, Bowen WD, Leonard ML (2009) Sex-specific, seasonal foraging tactics of adult grey seals (Halichoerus grypus) revealed by state space analysis. Ecology 90:3209–3221
Baum LE, Petrie T, Soules G, Weiss NA (1970) maximization technique occurring in the statistical analysis of probabilistic functions of Markov chains. Ann Math Stat 41:164–171
Bulla J, Lagona F, Maruotti A, Picone M (2012) A multivariate hidden Markov model for the identification of sea regimes from incomplete skewed and circular time series. J Agric Biol Environ Stat 17:544–567
Bulla J, Lagona F, Maruotti A, Picone M (2015) Environmental conditions in semi-enclosed basins: a dynamic latent class approach for mixed-type multivariate variables. Journal de la Societé Français de Statistique 156:114–136
Carnicero JA, Ausin MC, Wiper MP (2013) Non-parametric copulas for circularlinear and circularcircular data: an application to wind directions. Stoch Environ Res Risk Assess 27:1991–2002
D’Elia A (2001) A statistical model for orientation mechanism. Stat Methods Appl 10:157–174
Farcomeni A (2015) Generalized linear mixed models based on latent Markov heterogeneity structures. Scand J Stat 42:1127–1135
Fisher NI, Lee AJ (1992) Regression models for angular response. Biometrics 48:665–677
Gill G, Hangartner D (2010) Circular data in political science and how to handle it. Polit Anal 18:316–336
Hanks EM, Hooten MB, Johnson DS, Sterling JT (2011) Velocity-based movement modeling for individual and population level inference. PLoS One 6:e22795
Heiss F (2008) Sequential numerical integration in nonlinear state space models for microeconometric panel data. J Appl Econ 23:373–389
Hokimoto T, Kiyofuji H (2014) Effect of regime switching on behavior of albacore under the influence of phytoplankton concentration. Stoch Environ Res Risk Assess 28:1099–1124
Holzmann H, Munk A, Suster M, Zucchini W (2006) Hidden Markov models for circular and linear-circular time series. Environ Ecol Stat 13:325–347
Hornik K, Grün B (2014). movMF: an R package for fitting mixtures of von Mises–Fisher Distributions. J Stat Softw, vol 58
Jammalamadaka RA, SenGupta A (2001) Topics in circular statistics. World Scientific, Singapore
Johnson RA, Wehrly TE (1978) Some angular-linear distributions and related regression models. J Am Stat Assoc 73:602–606
Lagona F (2015) Regression analysis of correlated circular data based on the multivariate von Mises distribution. Environ Ecol Stat. doi:10.1007/s10651-015-0330-y
Lagona F, Jdanov D, Shkolnikova M (2014) Latent time-varying factor in longitudinal analysis: a linear mixed hidden Markov model for heart rates. Stat Med 33:4116–4134
Lagona F, Picone M, Maruotti A, Cosoli S (2015) A hidden Markov approach to the analysis of space–time environmental data with linear and circular components. Stoch Environ Res Risk Assess 29:397–409
Langrock R, King R, Matthiopoulos J, Thomas L, Fortin D, Morales JM (2012) Flexible and practical modeling of animal telemetry data: hidden Markov models and extensions. Ecology 93:2336–2342
Langrock R, Hopcraft JGC, Blackwell PG, Goodall V, King R, Niu M, Patterson TA, Pedersen MW, Skarin A, SchicK RS (2014a) Modelling group dynamic animal movement. Methods Ecol Evol 5:190–199
Langrock R, Marques TA, Baird RW, Thomas L (2014b) Modeling the diving behavior of whales: a latent-variable approach with feedback and semi-Markovian components. J Agric Biol Environ Stat 19:82–100
Lee A (2010) Circular data. Wiley Interdiscip Rev 2:477–486
Leroux BG, Puterman ML (1992) Maximum-Penalized-Likelihood estimation for independent and Markov dependent mixture models. Biometrics 48:545–558
Maruotti A (2011) Mixed hidden Markov models for longitudinal data: an overview. Int Stat Rev 79:427–454
Maruotti A, Rocci R (2012) A mixed non-homogeneous hidden Markov model for categorical data, with application to alcohol consumption. Stat Med 9:871–886
Mastrantonio G, Jona-Lasinio G, Maruotti A (2015) Bayesian hidden Markov modelling using circular-linear general projected normal distribution. Environmetrics 26:145–158
McClintock BT, King R, Thomas L, Matthiopoulos J, McConnell BJ, Morales JM (2012) A general discrete-time modeling framework for animal movement using multi-state random walks. Ecol Monogr 82:335–349
McKellar AE, Langrock R, Walters JR, Kesler DC (2015) Using mixed hidden Markov models to examine behavioral states in a cooperatively breeding bird. Behav Ecol 26:148–157
McLellan CR, Worton BJ, Deasy W, Birch ANE (2015) Modelling larval movement data from individual bioassays. Biom J 57(3):485–501
Nathan R, Getz WM, Revilla E, Holyoak M, Kadmon R, Saltz D, Smouse PE (2008) A movement ecology paradigm for unifying organismal movement research. Proc Natl Acad Sci 105:19052–19059
Nunez-Antonio G, Gutierrez-Pena E (2014) A Bayesian model for longitudinal circular data based on the projected normal distribution. Comput Stat Data Anal 71:506–519
Patlak CS (1953a) A mathematical contribution to the study of orientation of organisms. Bull Math Biophys 15:431–476
Patlak CS (1953b) Random walk with persistence and external bias. Bull Math Biophys 15:311–338
Patterson TA, Thomas L, Wilcox C, Ovaskainen O, Matthiopoulos J (2008) State-space models of individual animal movement. Trends Ecol Evol 23:87–94
Presnell B, Morrison SP, Littell RC (1998) Projected multivariate linear model for directional data. J Am Stat Assoc 93:1068–1077
Song PXK (2007) Correlated data analysis. Springer, Berlin
Visser I, Raijmakers M, Molenaar P (2000) Confidence intervals for hidden Markov model parameters. Br J Math Stat Psychol 53:317–327
Visser I, Raijmakers MEJ, Molenaar PCM (2002) Fitting hidden Markov models to psychological data. Sci Program 10:185–199
Wang F, Gelfand A (2013) Directional data analysis under the general projected normal distribution. Stat Methodol 10:113–127
Wang F, Gelfand A (2014) Modeling space and space–time directional data using projected Gaussian processes. J Am Stat Assoc 109:1565–1580
Wang F, Gelfand A, Jona-Lasinio G (2015) Joint spatio-temporal analysis of a linear and a directional variable: space–time modeling of wave heights and wave directions in the Adriatic Sea. Stat Sin 25:25–39
Author information
Authors and Affiliations
Corresponding author
Appendices
Appendix 1: The mixed projected normal model
The mixed project normal model
relaxes the independence assumption on repeated measurements and may account for correlations between projections. Indeed, by assuming that projections are conditionally independent given the covariates and the random effects, for the i-th unit, the likelihood contribution would be
where \(g(\cdot )\) is the random effects density function. Nevertheless, in practice, \(g(b_{i1},b_{i2}) = g_1(b_{i1})g_2(b_{i2})\), i.e. random effects are assumed independent and, accordingly, projections can be separately modelled and are independent as well. Thus, although theoretically the mixed projected normal model could allow for conditional independence, this feature is not investigated or modelled in the literature. Formally, if \(g(b_{i1},b_{i2}) = g_1(b_{i1})g_2(b_{i2})\), we have
and, thus, the projections are independent.
Appendix 2: Computational details
As it stands the likelihood function of the hidden Markov projected normal model is of little or no computational use, because it involves a sum over \(K^T\) terms for each unit i and cannot be directly evaluated. It quickly becomes infeasible to compute even for small values of K as T grows to moderate size. Clearly, a more efficient procedure is needed to perform the calculation of the likelihood function. This issue may be addressed via the so-called forward variables (Baum et al. 1970). Let us start defining
which represents the probability of seeing the partial sequence ending up in state k at time t for a generic unit i. We can compute \(\alpha _{itk}\) recursively by
As a by-product of the forward procedure we find that the likelihood can be written as
Let us further define
i.e. the probability of the partial sequence \(({\mathbf{y}}_{it+1},\ldots ,{\mathbf{y}}_{iT})\) given that the i-th unit started in state k at time t. The backward recursion is given by
We can express \(\hat{\xi }_{itk}\) and \(\hat{\zeta }_{itjk}\) in terms of forward and backward variables by
At last, we have
where \({\varvec{\mu }}_{itk} = (\mu _{it1k},\mu _{it2k})\) is the state-specific vector of projections’ means and \({\varvec{{{\varPhi}}}} (\cdot )\) is the cdf of a bivariate standard normal distribution.
Rights and permissions
About this article
Cite this article
Maruotti, A., Punzo, A., Mastrantonio, G. et al. A time-dependent extension of the projected normal regression model for longitudinal circular data based on a hidden Markov heterogeneity structure. Stoch Environ Res Risk Assess 30, 1725–1740 (2016). https://doi.org/10.1007/s00477-015-1183-5
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00477-015-1183-5