Abstract
Main conclusion
Two TFL1 -like genes, CorfloTFL1 and CorcanTFL1 cloned from Cornus florida and C. canadensis, function in regulating the transition to reproductive development in Arabidopsis.
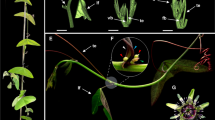
TERMINAL FLOWER 1 (TFL1) is known to regulate inflorescence development in Arabidopsis thaliana and to inhibit the transition from a vegetative to reproductive phase within the shoot apical meristem. Despite the importance, TFL1 homologs have been functionally characterized in only a handful eudicots. Here we report the role of TFL1 homologs of Cornus L. in asterid clade of eudicots. Two TFL1-like genes, CorfloTFL1 and CorcanTFL1, were cloned from Cornus florida (a tree) and C. canadensis (a subshrub), respectively. Both are deduced to encode proteins of 175 amino acids. The amino acid sequences of these two Cornus TFL1 homologs share a high similarity to Arabidopsis TFL1 and phylogenetically more close to TFL1 paralogous copy ATC (Arabidopsis thaliana CENTRORADIALIS homologue). Two genes are overexpressed in wild-type and tfl1 mutant plants of A. thaliana. The over-expression of each gene in wild-type Arabidopsis plants results in delaying flowering time, increase of plant height and cauline and rosette leaf numbers, excessive shoot buds, and secondary inflorescence branches. The over-expression of each gene in the tfl1 mutant rescued developmental defects, such as the early determinate inflorescence development, early flowering time, and other vegetative growth defects, to normal phenotypes of wild-type plants. These transgenic phenotypes are inherited in progenies. All data indicate that CorfloTFL1 and CorcanTFL1 have conserved the ancestral function of TFL1 and CEN regulating flowering time and inflorescence determinacy.






Similar content being viewed by others
References
Abe M, Kobayashi Y, Yamamoto S, Daimon Y, Yamaguchi A, Ikeda Y, Ichinoki H, Notaguchi M, Goto K, Araki T (2005) FD, a bZIP protein mediating signals from the floral pathway integrator FT at the shoot apex. Science 309(5737):1052–1056
Ahn JH, Miller D, Winter VJ, Banfield MJ, Lee JH, Yoo SY, Henz SR, Brady RL, Weigel D (2006) A divergent external loop confers antagonistic activity on floral regulators FT and TFL1. The EMBO Journal 25(3):605–614
Amaya I, Ratcliffe OJ, Bradley DJ (1999) Expression of CENTRORADIALIS (CEN) and CEN-like genes in tobacco reveals a conserved mechanism controlling phase change in diverse species. The Plant Cell Online 11(8):1405–1417
Boss PK, Bastow RM, Mylne JS, Dean C (2004) Multiple pathways in the decision to flower: enabling, promoting, and resetting. Plant Cell Online 16(suppl 1):S18–S31
Bradley D, Carpenter R, Copsey L, Vincent C, Rothstein S, Coen E (1996) Control of inflorescence architecture in Antirrhinum. Nature 379:791–797
Bremer B, Bremer K, Chase M, Fay M, Reveal J, Soltis D, Soltis P, Stevens P (2009) An update of the Angiosperm phylogeny group classification for the orders and families of flowering plants: APG III. Bot J Linnean Soc 161(2):105–121
Chang S, Puryear J, Cairney J (1993) A simple and efficient method for isolating RNA from pine trees. Plant Mol Biol Rep 11(2):113–116
Chardon F, Damerval C (2005) Phylogenomic analysis of the PEBP gene family in cereals. J Mol Evol 61(5):579–590
Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16(6):735–743
Conti L, Bradley D (2007) TERMINAL FLOWER1 is a mobile signal controlling Arabidopsis architecture. Plant Cell Online 19(3):767–778
Danilevskaya ON, Meng X, Hou Z, Ananiev EV, Simmons CR (2008) A genomic and expression compendium of the expanded PEBP gene family from maize. Plant Physiol 146(1):250–264
Danilevskaya ON, Meng X, Ananiev EV (2010) Concerted modification of flowering time and inflorescence architecture by ectopic expression of TFL1-like genes in maize. Plant Physiol 153(1):238–251
Espinosa-Soto C, Padilla-Longoria P, Alvarez-Buylla ER (2004) A gene regulatory network model for cell-fate determination during Arabidopsis thaliana flower development that is robust and recovers experimental gene expression profiles. Plant Cell Online 16(11):2923–2939
Feng C-M, Qu R, Zhou L-L, Xie D-Y, Xiang Q-YJ (2009) Shoot regeneration of dwarf dogwood (Cornus canadensis L.) and morphological characterization of the regenerated plants. Plant Cell Tissue Organ Culture (PCTOC) 97(1):27–37
Feng CM, Xiang QYJ, Franks RG (2011) Phylogeny-based developmental analyses illuminate evolution of inflorescence architectures in dogwoods (Cornus sl, Cornaceae). New Phytol 191(3):850–869
Feng C-M, Liu X, Yu Y, Xie D, Franks RG, Xiang Q-YJ (2012) Evolution of bract development and B-class MADS box gene expression in petaloid bracts of Cornuss. l. (Cornaceae). New Phytol 196:1–13
Hake S (2008) Inflorescence architecture: the transition from branches to flowers. Curr Biol 18(23):R1106–R1108. doi:10.1016/j.cub.2008.10.024
Hanzawa Y, Money T, Bradley D (2005) A single amino acid converts a repressor to an activator of flowering. Proc Natl Acad Sci USA 102(21):7748–7753
Harrison SJ, Mott EK, Parsley K, Aspinall S, Gray JC, Cottage A (2006) A rapid and robust method of identifying transformed Arabidopsis thaliana seedlings following floral dip transformation. Plant Methods 2(1):19
Hedman H, Källman T, Lagercrantz U (2009) Early evolution of the MFT-like gene family in plants. Plant Mol Biol 70(4):359–369
Ho WWH, Weigel D (2014) Structural features determining flower-promoting activity of Arabidopsis FLOWERING LOCUS T. The Plant Cell Online 26(2):552–564
Hofgen R, Willmitzer L (1988) Storage of competent cells for Agrobacterium transformation. Nucleic Acids Res 16(20):9877–9878
Kang S, Lee J, Lee MS, Seok C (2008) Structural basis of functional conversion of a floral repressor to an activator: a molecular dynamics simulation study. Bull-Korean Chem Soc 29(2):408
Karimi M, Inzé D, Depicker A (2002) GATEWAY™ vectors for Agrobacterium-mediated plant transformation. Trends Plant Sci 7(5):193–195
Karlgren A, Gyllenstrand N, Källman T, Sundström JF, Moore D, Lascoux M, Lagercrantz U (2011) Evolution of the PEBP gene family in plants: functional diversification in seed plant evolution. Plant Physiol 156(4):1967–1977
Karlgren A, Gyllenstrand N, Clapham D, Lagercrantz U (2013) FLOWERING LOCUS T/TERMINAL FLOWER1-like genes affect growth rhythm and bud set in Norway spruce. Plant Physiol 163(2):792–803
Kasahara T, Miyazaki T, Nitta H, Ono A, Miyagishima T, Nagao T, Urushidani T (2006) Evaluation of methods for duration of preservation of RNA quality in rat liver used for transcriptome analysis. J Toxicol Sci 31(5):509–519
Kotoda N, Wada M (2005) MdTFL1, a TFL1-like gene of apple, retards the transition from the vegetative to reproductive phase in transgenic Arabidopsis. Plant Sci 168(1):95–104
Li C, Luo L, Fu Q, Niu L, Xu Z-F (2015) Identification and characterization of the FT/TFL1 gene family in the biofuel plant Jatropha curcas. Plant Mol Biol Rep 33(2):326–333
Liljegren SJ, Gustafson-Brown C, Pinyopich A, Ditta GS, Yanofsky MF (1999) Interactions among APETALA1, LEAFY, and TERMINAL FLOWER1 specify meristem fate. Plant Cell Online 11(6):1007–1018
Lippman ZB, Cohen O, Alvarez JP, Abu-Abied M, Pekker I, Paran I, Eshed Y, Zamir D (2008) The making of a compound inflorescence in tomato and related nightshades. PLoS Biol 6(11):e288
Liu N, Sliwinski MK, Correa R, Baum DA (2011) Possible contributions of TERMINAL FLOWER 1 to the evolution of rosette flowering in Leavenworthia (Brassicaceae). New Phytol 189(2):616–628
Liu X, Feng C-M, Franks R, Qu R, Xie D-Y, Xiang Q-YJ (2013) Plant regeneration and genetic transformation of C. canadensis: a non-model plant appropriate for investigation of flower development in Cornus (Cornaceae). Plant cell reports 32(1):77–87
Miller MA, Pfeiffer W, Schwartz T (2010) Creating the CIPRES science gateway for inference of large phylogenetic trees. Proc Gateway Comput Environ Workshop New Orleans LA 14:1–8
Mimida N, Goto K, Kobayashi Y, Araki T, Ahn JH, Weigel D, Murata M, Motoyoshi F, Sakamoto W (2001) Functional divergence of the TFL1-like gene family in Arabidopsis revealed by characterization of a novel homologue. Genes Cells 6(4):327–336
Mimida N, Kotoda N, Ueda T, Igarashi M, Hatsuyama Y, Iwanami H, Moriya S, Abe K (2009) Four TFL1/CEN-like genes on distinct linkage groups show different expression patterns to regulate vegetative and reproductive development in apple (Malus × domestica Borkh.). Plant Cell Physiol 50(2):394–412
Mizukami Y, Ma H (1997) Determination of Arabidopsis floral meristem identity by AGAMOUS. Plant Cell 9:3393–3408
Mohamed R, Wang CT, Ma C, Shevchenko O, Dye SJ, Puzey JR, Etherington E, Sheng X, Meilan R, Strauss SH (2010) Populus CEN/TFL1 regulates first onset of flowering, axillary meristem identity and dormancy release in Populus. Plant J 62(4):674–688
Murashige T, Skoog F (1962) A revised medium for rapid growth and bio assays with tobacco tissue cultures. Physiol Plant 15(3):473–497
Nilsson O, Lee I, Blázquez MA, Weigel D (1998) Flowering-time genes modulate the response to LEAFY activity. Genetics 150(1):403–410
Ohshima S, Murata M, Sakamoto W, Ogura Y, Motoyoshi F (1997) Cloning and molecular analysis of the Arabidopsis gene Terminal Flower 1. Mol Gen Genet MGG 254(2):186–194
Périlleux C, Lobet G, Tocquin P (2014) Inflorescence development in tomato: gene functions within a zigzag model. Front Plant Sci 5:1–12
Pnueli L, Carmel-Goren L, Hareven D, Gutfinger T, Alvarez J, Ganal M, Zamir D, Lifschitz E (1998) The SELF-PRUNING gene of tomato regulates vegetative to reproductive switching of sympodial meristems and is the ortholog of CEN and TFL1. Development 125(11):1979–1989
Pnueli L, Gutfinger T, Hareven D, Ben-Naim O, Ron N, Adir N, Lifschitz E (2001) Tomato SP-interacting proteins define a conserved signaling system that regulates shoot architecture and flowering. Plant Cell Online 13(12):2687–2702
Posada D (2008) jModelTest: phylogenetic model averaging. Mol Biol Evol 25:1253–1256. doi:10.1093/molbev/msn083
Ratcliffe OJ, Amaya I, Vincent CA, Rothstein S, Carpenter R, Coen ES, Bradley DJ (1998) A common mechanism controls the life cycle and architecture of plants. Development 125(9):1609–1615
Repinski S, Kwak M, Gepts P (2012) The common bean growth habit gene PvTFL1y is a functional homolog of Arabidopsis TFL1. Theor Appl Genet 124(8):1539–1547
Ronquist F, Huelsenbeck JP (2003) Mrbayes 3: bayesian phylogenetic inference under mixed models. Bioinformatics 19:1572–1574
Ronquist F, Klopfstein S, Vilhelmsen L, Schulmeister S, Murray DL, and Rasnitsyn AP (2012) A total-evidence approach to dating, applied to the early radiation of hymenoptera. Syst Biol 61(6):973–999
Rosso MG, Li Y, Strizhov N, Reiss B, Dekker K, Weisshaar B (2003) An Arabidopsis thaliana T-DNA mutagenized population (GABI-Kat) for flanking sequence tag-based reverse genetics. Plant Mol Biol 53(1–2):247–259
Schultz EA, Haughn GW (1993) Genetic analysis of the floral initiation process (FLIP) in Arabidopsis. Development 119(3):745–765
Shannon S, Meeks-Wagner DR (1991) A mutation in the Arabidopsis TFL1 gene affects inflorescence meristem development. The Plant Cell Online 3(9):877–892
Wang R, Albani MC, Vincent C, Bergonzi S, Luan M, Bai Y, Kiefer C, Castillo R, Coupland G (2011) Aa TFL1 confers an age-dependent response to vernalization in perennial Arabis alpina. Plant Cell Online 23(4):1307–1321
Wellmer F, Riechmann JL (2010) Gene networks controlling the initiation of flower development. Trends Genet 26(12):519–527
Wigge PA, Kim MC, Jaeger KE, Busch W, Schmid M, Lohmann JU, Weigel D (2005) Integration of spatial and temporal information during floral induction in Arabidopsis. Science 309(5737):1056–1059
Xiang Q-Y, Thomas DT (2008) Tracking character evolution and biogeographic history through time in Cornaceae—does choice of methods matter. J System Evol 46(3):349–374
Xiang Q-Y, Thomas DT, Zhang W, Manchester SR, Murrell Z (2006) Species level phylogeny of the genus Cornus (Cornaceae) based on molecular and morphological evidence-implications for taxonomy and Tertiary intercontinental migration. Taxon 55(1):9–30
Zhang J, Franks RG, Liu X, Kang M, Keebler JE, Schaff JE, Huang H-W, Xiang Q-YJ (2013) De novo sequencing, characterization, and comparison of inflorescence transcriptomes of Cornus canadensis and C. florida (Cornaceae). PLoS One 8(12):e82674
Acknowledgements
We would like to thank NCSU Phytotron for culturing plants of C. canadensis and A. thaliana, JC Raulston Arboretum for providing Cornus plants as the sources for experimental materials. We are grateful to undergraduate students Charlotte Rastas, Chanel Wilson, Kimberly Shearer, and Ashley Yow for assistance in culturing the transgenic and control plants, Ashley Yow for DNA extraction and PCR of transgenic plants, and to the anonymous reviewers for critical review of the manuscript and constructive comments. The study was supported by a National Science Foundation of the United States grant (IOS-1024629).
Author information
Authors and Affiliations
Corresponding authors
Electronic supplementary material
Below is the link to the electronic supplementary material.
425_2016_2466_MOESM1_ESM.jpg
Phenotypes of T2 generation of transgenic plants. (a) Averages of T2 lines of 35S::CorcanTFL1 Col-0 transformation. (b) Averages of T2 lines of 35S::CorfloTFL1 Col-0 transformation. (c) Averages of T2 lines of CorcanTFL1 tfl1-18 transformation. (d) Averages of T2 lines of CorfloTFL1 tfl1-18 transformation. 15 plants of each line were planted. An average of 15 plants of each T1 offspring was calculated first. The average was then used for calculating the total average of all lines of each transgene (CorcanTFL1 or CorfloTFL1) with each genetic background (Col-0 or tfl1-18). Columns indicate the averages of each phenotype. Error bars indicate the standard deviation (±SD) (JPEG 289 kb)
Rights and permissions
About this article
Cite this article
Liu, X., Zhang, J., Abuahmad, A. et al. Analysis of two TFL1 homologs of dogwood species (Cornus L.) indicates functional conservation in control of transition to flowering. Planta 243, 1129–1141 (2016). https://doi.org/10.1007/s00425-016-2466-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00425-016-2466-x