Abstract
For proper mammalian brain development and functioning, the translation of many neuronal mRNAs needs to be repressed without neuronal activity stimulations. We have discovered that the expression of a subclass of neuronal proteins essential for neurodevelopment and neuron plasticity is co-regulated at the translational level by TDP-43 and the Fragile X Syndrome protein FMRP. Using molecular, cellular and imaging approaches, we show that these two RNA-binding proteins (RBP) co-repress the translation initiation of Rac1, Map1b and GluR1 mRNAs, and consequently the hippocampal spinogenesis. The co-repression occurs through binding of TDP-43 to mRNA(s) at specific UG/GU sequences and recruitment of the inhibitory CYFIP1-FMRP complex by its glycine-rich domain. This novel regulatory scenario could be utilized to silence a significant portion of around 160 common target mRNAs of the two RBPs. The study establishes a functional/physical partnership between FMRP and TDP-43 that mechanistically links several neurodevelopmental disorders and neurodegenerative diseases.
Similar content being viewed by others
Introduction
Neurodevelopmental disorders result from growth and developmental impairments of the brain or central nervous system. Cognitive and neuropsychiatric problems and intellectual disability in patients with these disorders are believed to be due to perturbations of synapse functioning with connectivity defects [44, 62]. Among the neurodevelopmental diseases, Fragile X Syndrome (FXS) is the most common inherited disorder, with intellectual impairment caused by the mutation of a single gene, FMR1, encoding the FMRP protein. Depletion of FMRP in diseased cells causes an increase in the translation of a number of proteins important for dendritic spine development [16, 59]. Of great genetic and clinical interest is the association of FMRP with other neurodevelopmental disorders, including autism spectrum disorders (ASD), as well as neurodegenerative diseases such as the fragile X-associated tremor ataxia syndrome (FXTAS) and Alzheimer’s disease (AD) [28, 65]. Significantly, while the genetic causes of the majority of ASD cases are unknown, ~5 % of ASD children also have FXS, with a mutated FMR1 gene and lower level of FMRP protein [22].
In contrast, neurodegenerative diseases are characterized by progressive loss of the structure/function of neurons and neuronal death at later stages in life. In neurodegenerative diseases such as Huntington’s disease (HD), Alzheimer’s disease (AD), frontotemporal lobar dementia (FTLD) and amyotrophic lateral sclerosis (ALS), abnormal protein-folding leads to aggregate formation, resulting in loss-of function of multiple proteins and giving rise to disease phenotypes [69]. Importantly, different RNA-binding proteins (RBPs) are involved extensively in various forms of neurodegenerative diseases and neurodevelopmental disorders [30, 58]. RBPs play important roles in RNA processing and metabolism, including pre-mRNA splicing, polyadenylation, transport, surveillance, mRNA localization, mRNA stability control, translational control and RNA editing. Aberrant expression and mutations in RBP genes affect various RNA processing steps and alter the target gene expression [25]. There are more than 800 RBPs encoded by the human genome and, together, they comprise approximately 40 different types of domain motifs [54]. Although considerable effort has been invested in understanding the biological functions of RBPs and the pathogenic mechanisms underlying RBP-associated diseases, a number of questions remain to be addressed.
TDP-43 is a ubiquitously expressed RNA-binding protein required for early development [68], and it has been implicated in multiple cellular processes including cell cycle progression, apoptosis, RNA processing, alternative splicing, etc., despite little being known about its associated mechanisms [4, 38, 42, 67]. TDP-43 binds at UG-rich sequence(s) on single-stranded RNAs with high affinity via its two RNA-binding domains, RRM1 and RRM2 [4]. Mis-metabolism of this protein, including the formation of cytoplasmic TDP-43(+) and ubiquitin(+) aggregates (TDP-43 proteinopathies), appears to be associated with the majority of ALS (ALS-TDP) and FTLD (FTLD-TDP) cases [29, 42, 49]. Numerous putative RNA substrates for TDP-43 binding, including TDP-43 mRNA itself, have been identified by global or targeted analyses which suggest its role in regulating the architecture and functions of neurons and synapses [52, 57, 61]. This is consistent with TDP-43 loss-of-neuronal-function being one of the major causes for the development of neurodegenerative diseases through TDP-43 proteinopathies [41, 69]. On the other hand, gain-of-toxicity has also been suggested to contribute to the pathogenesis of ALS-TDP and FTLD-TDP [8, 41, 63].
Several studies have suggested that one of the functions of TDP-43 in neurons is to regulate translation. First, TDP-43 represses translation in an in vitro system [66]. Second, TDP-43 interacts with translational regulators, including FMRP and several heterogeneous ribonucleoprotein particles (hnRNPs) [24]. Third, in a Drosophila ALS model with over-expression of human TDP-43 in the motor neurons, translation of futsch (the Drosophila ortholog of human Map1b) appears to be impaired [14] and this impairment is partially rescued by overexpression of FMRP [15]. Finally, expression of one of the positive regulators of spinogenesis, Rac1, appears to be controlled by TDP-43 at the level of translation [45]. Despite these indications, whether TDP-43 directly regulates translation in neurons in vivo is still unclear and, if it does, the mechanistic details of TDP-43-mediated translational regulation are unknown.
Unlike TDP-43, FMRP is a well-established repressor of translation and it acts by blocking either the initiation or elongation step [12]. Approximately 4 % of mouse brain mRNAs interact with FMRP. Furthermore, FMRP can directly bind mRNAs that possess the G-quadruplex structure through one of its three RNA-binding domains [46, 64]. It has also been suggested that the association between FMRP and mRNAs may require yet to be identified adaptor proteins [20, 48] or RNAs such as BC1 [72]. In an FXS mouse model, loss of FMRP causes an increased global level of protein synthesis [50], resulting in a high density of dendritic spines [10] as well as affecting the brain development and synaptic plasticity [59]. Notably, the increased dendritic spine density and likely the FXS pathology could be attributed in part to an increase of Rac1 protein expression due to the loss of FMRP [6]. Furthermore, FMRP is associated with Rac1 mRNA in ribonucleoprotein (RNP) granules [40]. This is in interesting parallel to the suggested regulatory role of TDP-43 in the translation of Rac1 mRNA and spinogenesis in hippocampal neurons, as revealed by the change in Rac1 protein amount, but not Rac1 mRNA levels and/or Rac1 protein stability, upon over-expression or knockdown of TDP-43 [45].
Here, we show for the first time a functional and mechanistic link between TDP-43 and FMRP in the translational regulation of several FMRP target mRNAs important for synaptic plasticity. Using Rac1 mRNA as the paradigm, we demonstrate that TDP-43 acts as an adaptor protein to recruit the FMRP-CYFIP1 inhibitory complex to mRNAs, thereby repressing the initiation of translation. This is an important advancement towards understanding the molecular mechanisms of both RBPs in translational regulation and unraveling the overlap between FMRP-associated neurodevelopmental disorders and neurodegenerative diseases at the molecular level.
Materials and methods
Primary mouse hippocampal neuronal culture and HEK293T cell culture
The 14 day pregnant FVB mice were obtained from the National Laboratory Animal Center of Taiwan. The preparation of primary hippocampal neurons in culture followed the standard protocols using cells mechanically dissociated from the hippocampi of E16.5–17.5 mouse embryos [45]. For the study of translational repression, cultured neurons were treated with the translational inhibitor 4EGI-1(Merck Millipore, Germany) at a final concentration of 25 μM for 30 min to 1 h before harvesting for further analysis.
Human HEK293T cell culture was grown at 37 °C in DMEM containing 10 % fetal bovine serum, 100 U/ml penicillin, and 100 g/ml streptomycin.
Plasmid construction
See Supplementary Experimental Procedure for detailed description of the different plasmids used in the study.
Transfection of plasmid DNAs and RNAi oligos
For details, see Supplementary Experimental Procedures.
Western blotting and RT-PCR analysis
For details, see Supplementary Experimental Procedures.
Immunofluorescence staining
Details of the immunofluorescence staining experiments of the primary hippocampal neurons are described in Supplementary Experimental Procedures.
Fluorescence in situ hybridization (FISH) and combined immunofluorescence (IF) staining
For details, see Supplementary Experimental Procedures.
Polysome profile analysis
The analysis followed the general procedures [39]. For details, see Supplementary Experimental Procedures.
RNA-IP
RNA-IP was used to study the protein-RNA interactions. For details see Supplementary Experimental Procedures.
Immunoprecipitation (IP)
For details, see Supplementary Experimental Procedures.
In vitro RNA–protein binding assay using biotinylated oligoneucleotides
The procedures followed those described before [7]. For details see Supplementary Experimental Procedures.
Dual-luciferase reporter assay of translation
For details, see Supplemental Experimentary Procedures.
Local dendritic translation assay
The details of the analysis by a reporter assay are described in Supplementary Experimental Procedures.
Results
Regulation of Rac1 mRNA translation by TDP-43 and FMRP in primary hippocampal neurons and non-neuronal cells
To unequivocally demonstrate that TDP-43 and FMRP regulate Rac1 mRNA translation, we transfected DIV 6 primary hippocampal neurons with different RNAi oligos including control Sc oligo, TDP-si oligo or FMRP-si oligo ([45, 53]; see Supplementary Materials and Methods for more details). As expected from previous studies [6, 45], Western blotting showed a significant increase in Rac1 protein levels upon depletion of either TDP-43 or FMRP by RNAi (Fig S1a). We then compared the polysome profiles of the cytoplasmic extracts from the different RNAi oligo-transfected primary hippocampal neurons by sucrose gradient sedimentation analysis. The polysome profiles of DIV 6–8 primary neurons, as exemplified on top of Fig. 1a(i), were similar to those of DIV 4–7 primary cortical neurons [34]. Significantly, while depletion of either TDP-43 or FMRP did not impair the global cellular translation status as reflected by the polysome profiles (data not shown), polysomal accumulation of Rac1 mRNA, but not of Gapdh mRNA, at the expense of its association with 40S and 60S/80S monosomes was evident in TDP-si or FMRP-si transfected cells [results from three biological repeats have been presented in Fig. 1a(ii) and results of three technical repeats from single biological sample was exemplified in Fig S1b]. Repression of Rac1 translation by TDP-43 and by FMRP in the primary hippocampal neurons, as reflected by the polysome profile analysis, was consistent with the [35S]-metabolic labeling and IP experiments (Fig S1c).
Effects of depletion of TDP-43/FMRP or over-expression of TDP-43 on the polysomal distribution of Rac1 mRNA. a Polysomal distribution of different proteins (i) and mRNAs (ii) in mouse primary hippocampal neurons. Cytoplasmic extracts, from cultured DIV 6 neurons transfected with different siRNA oligos, were separated by sucrose gradient sedimentation. i Top, representative polysome profile of DIV 6 primary hippocampal neurons. Bottom, different fractions of the sucrose gradient loaded with extract from Sc oligo-transfected hippocampal neurons were analyzed by Western blotting using different antibodies. ii Quantitative RT-PCR analysis of Rac1 mRNA in 40S monosome (left histogram), 60S/80S monosomes (middle histogram), and polysome (right histogram) fractions of DIV 6 primary hippocampal neurons transfected with Sc, TDP-si and FMRP-si oligos. The experiments were repeated 3 times for each of three different preparations of neuron culture (N = 3). Statistical significance between control Sc oligo and TDP-si or FMRP-si oligo transfected neurons was determined by Student’s t test: ***p < 0.0001 and **p < 0.001. Inputs were similar (data not shown). The graphs of polysomal distribution from use of one of the three biological samples are exemplified in Fig S1b. b Polysomal distributions of Rac1 mRNAs in HEK293T cells with ectopic expression of TDP-43 and/or depletion of FMRP by FMRP-si oligo. i Top, Representative polysomal profile of HEK293T cells. Bottom, Western blot analysis of the distribution patterns of different proteins in the polysomal fractions of HEK293T cells over-expressing pFlag-TDP-43. ii Quantitative RT-PCR analysis of Rac1 mRNA in 40S monosome (left histogram), 60S/80S monosomes (middle histogram), and polysome (right histogram) fractions of HEK293T cells transfected with pFlag, pFlag-TDP-43 ± FMRP-si oligo. The experiments were repeated 3 times for each of three different sets of extract preparation (N = 3). Significant differences among the three groups were determined by one way ANOVA, ***p < 0.0001. Inputs were similar (data not shown). The graphs of polysomal distribution from use of one of the three biological samples are exemplified in Fig S1d
We also examined the distributions of TDP-43 and FMRP proteins in the sucrose gradients. As shown by Western blotting [bottom panel, Fig. 1a(i)], both proteins were co-fractionated mainly with mRNPs, 40S, and partially with 60S/80S. Notably, the distribution of FMRP-containing RNP complexes in sucrose gradients has been studied by many groups because of its important role in translation. Surprisingly, different groups have found prominent differences in the distribution patterns of FMRP in sucrose gradient fractions. It was reported to be co-sedimenting with the translating polyribosomes [17, 33], equally distributed between the polysomes or monosomes and mRNPs [5, 9, 27], or mostly non-polysomal and associated with the translation initiation complexes [48, 71]. These discrepancies could be caused in part by differences in the extract preparation procedures and/or the different sources from which the extracts were prepared [60]. In any case, as shown in Fig. 1a(i), we found that distribution pattern of FMRP protein in the polysome profile of DIV 6-8 primary hippocampal neuron extract was different from that of the ribosomal protein rpL6 but similar to those of the translational regulators eIF4E and CYFIP1, which were known to interact with FMRP to inhibit translation at the initiation step [48]. Thus, our results indicated that FMRP in the hippocampal neurons might function in the inhibition of the translation initiation as reported before for the synaptoneurosomes [48]. The similar distribution patterns of TDP-43 and FMRP further suggested that TDP-43 might reside within the same complex(es) with FMRP, CYFIP1, eIF4E, and perhaps also with some of the translationally silent mRNAs.
The polysome profiles of HEK293T cells, as exemplified in Fig. 1b(i), were similar with or without exogenous expression of Flag-TDP-43 or depletion of endogenous TDP-43 or FMRP by respective RNAi (data not shown). Notably, the polysome profiles and FMRP distribution pattern shown in Fig. 1b(i) were similar to those of the primary hippocampal neurons [Fig. 1a(i)] and lymphoblast cell lines [9]. Interestingly, the exogenous Flag-TDP-43 partially co-sedimented with FMRP and CYFIP1 [bottom panel, Fig. 1b(i)], suggesting that exogenous TDP-43 resided in the translation inhibitory complexes containing the endogenous FMRP and CYFIP1. Furthermore, the level of Rac1 mRNA, but not Gapdh mRNA, in the monosomal fractions was increased with a concomitant decrease in the polysomal fractions upon over-expression of Flag-TDP-43 (results from three biological repeats have been presented in Fig. 1b(ii) and results of three technical repeats from single biological sample was exemplified in Fig S1d). Finally, it was interesting to note that, the decrease of Rac1 mRNA in the polysomal fractions could be partially rescued by depletion of FMRP using the FMRP-si oligo [Fig. 1b(ii)]. Together, these data suggest that TDP-43 and FMRP might reside in the same mRNP complexes and that TDP-43 inhibits Rac1 mRNA translation in a FMRP-dependent manner.
Physical interaction of TDP-43 with Rac1 mRNA and FMRP
To further understand the regulation of Rac1 mRNA translation by TDP-43 and FMRP, we first tested whether TDP-43 and FMRP could form an RNP complex together with Rac1 mRNA. RNA-immunoprecipitation assays (IP) of primary hippocampal neuronal cell extracts were carried out using anti-TDP-43 and anti-FMRP under conditions that preserved native protein-RNA complexes. The levels of Rac1 mRNA in the immunoprecipitated complexes were compared by quantitative [Fig. 2a(i)] and semi-quantitative [Fig. 2a(ii)] RT-PCR. In comparison to the control IgG, Rac1 mRNA was significantly enriched in the anti-TDP-43 or anti-FMRP immunoprecipitated complexes. However, the binding of FMRP to Rac1 mRNA was significantly decreased upon TDP-43 depletion, whereas only marginal or no decrease in the binding of TDP-43 was detected upon FMRP depletion [Fig. 2a(i)]. Data from the semi-quantitative RT-PCR analysis were consistent with the results of the quantitative RT-PCR analysis [compare lanes 4–6, Fig. 2a(ii)]. Thus, the binding of TDP-43 to Rac1 mRNA was independent of FMRP, whereas FMRP was recruited to Rac1 mRNA-containing RNPs in a TDP-43-dependent manner. Consistent with the above, Western blot analysis showed that proteins isolated from RNA-IP complexes pulled-down by anti-TDP-43 contained TDP-43 as well as FMRP (compare lane 4 with other lanes, Fig. 2b).
TDP-43 associated recruitment of FMRP in mRNP complex containing Rac1 mRNA in vivo. a RNA-IP. Total cellular extracts of cultured hippocampal neurons transfected with Sc, TDP-si or FMRP-si were immunoprecipitated with different antibodies, and the immunoprecipitated RNAs were analyzed (N = 3) by quantitative (i), presented as fold enrichment normalized to Sc-IgG, or semi-quantitative (ii) RT-PCR using primers specific for Rac1 mRNA. Hdac6 and PSD-95(Dlg4) mRNAs were also analyzed as positive controls and Gapdh mRNA as the negative control. Inputs were similar. Data in (i) are presented as mean ± SD, from three independent experiments. Significant changes in enrichment of Rac1 mRNA between IgG pulled down and anti-TDP-43 or anti-FMRP pulled down samples as well as between control Sc oligo and TDP-si or FMRP-si oligo transfected neurons are represented by ***p < 0.0001 (Student’s t test). b Identification of proteins associated with Rac1 mRNA. Proteins isolated from RNA-IP complexes were analyzed by Western blotting using anti-TDP-43 and anti-FMRP. The experiment was repeated several times
To investigate whether TDP-43 directly interacted with Rac1 mRNA, we used an in vitro transcription reaction protocol to generate biotin-labeled RNA probes corresponding to the 5′-untranslated region (5′UTR, probe A), the coding sequence (CDS, probe B), and two parts of the 3′-untranslated region (3′UTR, probes C and D) of Rac1 mRNA. We also generated three RNA probes from 3′UTR (C), namely 3′UTR-1, 3′UTR-2, 3′UTR-3, as well as mutated forms of these RNA probes harboring base changes to disrupt the putative TDP-43-binding UG/GU repeats, namely 3′UTR-2-Mt, 3′UTR-3-Mt, 3′UTR-3-Mt1 and CDS-Mt [left map, Fig. 3a(i)]. Purified probes were incubated with avidin-conjugated beads and extracts from HEK293T cells expressing Flag-TDP-43. As seen, TDP-43 could be pulled down by probes B, C [right panels of Fig. 3a(i)], CDS, 3′UTR-3 and 3′UTR-2 [lanes 1, 3 and 6, Fig. 3a(ii)], but not by 5′UTR (A), 3′UTR-4(D) [right panels of Fig. 3a(i)] or 3′UTR-1(data not shown). As expected, similar pull-down experiments demonstrated that both RNA-binding motifs of TDP-43 were required for effective binding with Rac1 mRNA [Fig. 3a(iii)]. In parallel, interactions between TDP-43 and 3′UTR-2, 3′UTR-3 or CDS were mostly abolished by the 3′UTR-2-Mt, 3′UTR-3-Mt and CDS-Mt mutations, respectively [lanes 2,5 and 7, upper panel of Fig. 3a(ii)], but not by 3′UTR-3-Mt1 [lane 4, upper panel of Fig. 3a(ii)]. Significantly, Western blotting analysis of the RNA probe-pulled-down samples using anti-FMRP revealed that RNA probes 3′UTR-2, 3′UTR-3 and mutated probe 3′UTR-3-Mt1, but not 3′UTR-2-Mt, 3′UTR-3-Mt, CDS and CDS-Mt, could efficiently pull-down endogenous FMRP [middle panel of Fig. 3a(ii)]. Whereas more structural studies would be needed to fully understand the nature of the physical interaction of TDP-43 and FMRP protein with Rac1 mRNA, the data of Figs. 2 and 3a together demonstrate that TDP-43 can bind to the 3′UTR and coding sequence of Rac1 mRNA at specific UG/GU dispersed repeats, but FMRP can only be associated with the 3′UTR of Rac1 mRNA and that this association is most likely mediated through recruitment of FMRP by the 3′UTR-bound TDP-43.
Binding and translational repression of Rac1 mRNA by TDP-43 and FMRP. a In vitro binding assay of TDP-43 to wild type and mutated Rac1 mRNA fragments. Biotinylated RNA probes corresponding to different parts of Rac1 mRNA, with or without base substitutions disrupting the putative TDP-43-binding UG/GU sequences (left map in i), were incubated with total cell extracts from HEK293T cells over-expressing Flag-TDP-43 (i, ii) and/or different deletion mutants of Flag-TDP-43 (iii). Affinity-purified elutes were separated on SDS-PAGE and immunoblotted with anti-Flag) top panels in i, ii, iii) or anti-FMRP (middle panel in ii). As the loading control, RNAs immunoprecipitated by anti-FLAG were also analyzed by semi-quantitative RT-PCR using primer set(s) specific for each of the corresponding RNA probes (bottom panels in i, ii, iii). All the experiments were repeated at least three times. b Luciferase reporter assay of HEK293T cells co-transfected with pFlag-TDP-43 and the psicheck2 reporter carrying full length wild type/mutant 3′UTR sequences of Rac1. Luciferase protein levels in cell extracts prepared at 48 h post-transfection were determined and presented as fold changes (N = 3), comparing cells co-expressing Flag-TDP-43 relative to cells co-expressing Flag. Significant differences in luciferase activity between Flag and Flag-TDP-43 transfected cells are represented by ***p < 0.0001 and *p < 0.01 (Student’s t test). c Luciferase activity assay of HEK293T cells co-transfected with the reporter containing Rac1 3′UTR, Sc oligo or FMRP-si oligo, and one or more of the following expression plasmids: pFlag, pGFP, pFlag-TDP-43 pGFP-FMRP, pFlag-TDP-43(ΔGly). Luciferase activities (N = 3) are represented as the fold change compared to cells transfected with Sc oligo, pFlag and pGFP. Rescue1 and Rescue 2 represent significant recovery of Rac1-3′UTR-mediated luciferase reporter translation. Significant changes are represented by ***p < 0.0001 and **p < 0.001 (Student’s t test). The amounts of RNA transcripts from different expression plasmids were similar in b and c (data not shown)
Translational co-repression of Rac1 mRNA mediated by TDP-43 and FMRP
Luciferase reporter assays were used to map the regions of Rac1 mRNA required for translational repression by TDP-43. 5′UTR, CDS and different 3′UTR fragments or their mutant counterparts were cloned into the dual luciferase vector psicheck2 and co-transfected with pFlag or pFlag-TDP-43 into HEK293T cells. Since the amounts of luciferase mRNAs were similar among different sets of transfected HEK293T cells [Fig S2a], any change in relative luciferase activity would occur at the translational level. As shown in Fig S2b, Flag-TDP-43 inhibited translation of the reporter RNAs containing 3′UTR-2 and 3′UTR-3 by 40 and 30 %, respectively, whereas the translation of the reporter RNAs containing 3′UTR-1, 3′UTR-4, 5′UTR or CDS were not inhibited. This data suggests that TDP-43 bound to the 3′UTR of Rac1 mRNA represses translation, but that the complex formed at the CDS harboring Flag-TDP-43 had no effect on translation. Thus, we also analyzed translational repression of the full-length 3′UTR of Rac1 by a luciferase reporter assay. As shown in Fig. 3b, translation of reporter RNA comprising the full length 3′UTR of Rac1 was halved by exogenous Flag-TDP-43. This repression could be rescued by the co-presence of 3′UTR-2-Mt and 3′UTR-3-Mt mutations (Fig. 3b).
The apparent requirement of FMRP for Flag-TDP-43-mediated translational repression of Rac1 mRNA (Fig. 1) was further established by luciferase reporter assays. First, siRNA-driven depletion of endogenous FMRP partially rescued (Rescue 1) translational repression of the luciferase reporter by Flag-TDP-43 and the 3′UTR of Rac1 mRNA [compare the 2nd and 3rd bars from left, Fig. 3c]. However, while co-expression of GFP-FMRP enhanced the extent of translational repression of the reporter by Flag-TDP-43 [compare 2nd and 4th bars from left, Fig. 3c], neither the exogenous Flag-TDP-43(ΔGly) that lacked the glycine-rich domain of TDP-43 nor Flag-TDP-43(ΔGly) + GFP-FMRP (Rescue 2) repressed translation (compare 4th bar with 5th and 6th bars from left, Fig. 3c). Notably, the glycine-rich domain of TDP-43 was required for its physical interaction with FMRP (compare lanes 1 and 2, Fig S3a). That GFP-FMRP alone could not significantly inhibit reporter RNA translation (compare the right-most bar to the left-most bar, Fig. 3c) might be due to the low level of endogenous TDP-43 in HEK293T cells [Fig S3b]. For similar reason, TDP-43 depletion by RNAi did not change the luciferase activity in HEK293T cells in comparison to the control cells (data not shown). These results demonstrate that FMRP and TDP-43 cooperatively function to repress translation of Rac1 mRNA through direct binding of TDP-43 to the 3′UTR region and physical interaction between the two RNA-binding factors.
Repression of translation initiation of Rac1 mRNA by TDP-43 through the FMRP-CYFIP1-mediated pathway
To investigate the mechanisms of translational co-repression of Rac1 mRNA by TDP-43 and FMRP, we first examined the effects of a selective inhibitor of eIF4E-eIF4G interactions (4EGI-1, [56]), on translation in primary hippocampal neurons. As expected, treatment of cultured neurons with 4EGI-1 led to a marked decrease in polysomal RNA content, indicative of global cellular translational inhibition [Fig. 4a(i)], as well as decreased level of Rac1 protein (data not shown). Similar to untreated primary hippocampal neurons (Fig. 1a), prior depletion of TDP-43 or FMRP by RNAi had little effect on the global polysome profile of 4EGI-1 treated cells (data not shown). However, the distribution of Rac1 mRNA (results of three technical repeats from single biological sample was exemplified in Fig S4a), but not Gapdh mRNA (results of three technical repeats from single biological sample was exemplified in Fig S4b), was significantly shifted from 40S and 60S/80S fractions to polysome fractions by treatment with TDP-si or FMRP-si oligo [results from three biological samples have been presented in Fig. 4a(ii)]. This shift was accompanied by increased levels of Rac1 protein, but not Gapdh, in the RNAi-treated neurons (data not shown). These data suggest that inhibition of Rac1 translation by 4EGI-1 at the initiation step could be selectively and partially rescued by depletion of TDP-43 or FMRP before 4EGI-1 treatment.
Translational co-repression of Rac1 mRNA by TDP-43 and FMRP at the initiation stage. a Effects of 4EGI-1, TDP-si oligo and FMRP-si oligo on polysomal distributions of Rac1 mRNA. i Representative polysome profiles of cultured DIV 6 hippocampal neurons with or without treatment of 25 µM 4EGI-1 for 30 min. ii Quantitative RT-PCR analysis of Rac1 mRNA in 40S monosome (left histogram), 60S/80S monosomes (middle histogram), and polysome (right histogram) fractions of DIV 6 primary hippocampal neurons transfected with Sc, TDP-si or FMRP-si oligo and with (represented as ‘4EGI-1’) or without (represented as ‘Mock’) treatment of 25 µM 4EGI-1 for 30 min. The results are presented by the histobar diagrams representing data from three different preparations of primary neuron culture (N = 3), each with three technical repeats of experiments. Significant differences between control Sc oligo and TDP-si or FMRP-si oligo transfected neurons as well as between Mock and 4EGI-1 treated neurons are represented by ***p < 0.0001 and **p < 0.001 (Student’s t test). Inputs were similar (data not shown). The graphs of polysomal distribution from use of one of the three biological samples are exemplified in Fig S4. b Luciferase reporter assay of HEK293T cells transfected with psicheck2-3′UTR reporter with (pFlag + pGFP), (pFlag-TDP-43 + pGFP) or (pFlag-TDP-43 + pGFP-FMRP), followed by treatment with 5 µM cyclohexamide (CHX) or 50 µM 4EGI-1 (4EGI-1) for 12 h. Luciferase activities are presented as fold changes relative to that of cells transfected with (pFlag + pGFP) and without any drug treatment (N = 3). Significant differences among different groups were determined by one way ANOVA, *p < 0.01. c IP analysis of the association of CYFIP1 with TDP-43, FMRP and Rac1 mRNA in DIV 6 primary hippocampal neurons. Cell lysates from primary hippocampal neurons transfected with siRNA oligos were immunoprecipitated with anti-CYFIP1 or anti-rabbit IgG, and then analyzed by Western blotting (top 4 panels on the right) or by RT-PCR (bottom 2 panels on the right). The experiment was repeated for 3 times with the statistical analysis shown in Fig S6b. Inputs were similar, as shown in the left lower 5 panels
Repression of translation by FMRP operates at the initiation step [48] or elongation step [17], depending on the targets and the cellular context. Therefore, we compared the effects of the elongation inhibitor cyclohexamide (CHX) and the initiation inhibitor 4EGI-1 on translational repression of the luciferase reporter psicheck2 carrying the 3′UTR of Rac1 mRNA by exogenous TDP-43 and/or FMRP in HEK293T cells. As seen in Fig. 4b, Flag-TDP-43 alone or Flag-TDP-43 + GFP-FMRP suppressed luciferase RNA translation even when elongation was blocked by CHX, indicating that elongation is not the rate limiting step for TDP-43/FMRP-mediated translational repression of Rac1 mRNA. However, cooperative translational repression was no longer evident when translation initiation was inhibited by 4EGI-1. Thus, the rate-limiting step of TDP-43/FMRP-targeted translational repression of Rac1 mRNA is indeed at the initiation step.
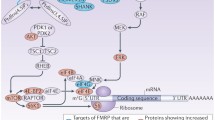
FMRP exerts its inhibitory role on translation initiation through the eIF4E-binding protein CYFIP1 [48]. Indeed, polysome profile analyses of primary hippocampal neurons [Fig. 1a(i)] and HEK293T cells [Fig. 1b(i)] showed partial co-sedimentation of TDP-43 and FMRP with CYFIP1, suggesting that they might form a complex together with mRNAs in vivo. To check whether CYFIP1 played a role in TDP-43/FMRP-mediated translational inhibition of Rac1 mRNA, we carried out RNA-IP analysis of extracts from primary hippocampal neurons transfected with control Sc oligo, FMRP-si oligo or TDP-si oligo using anti-CYFIP1. Significantly, CYFIP1 formed complexes with Rac1 mRNA, TDP-43, FMRP, as well as with eIF4E proteins (lane 1, Fig. 4c). However, the amount of Rac1 mRNA, as measured by RT-PCR, in the CYFIP1-pulled down RNP complex(s) decreased upon depletion of either FMRP or TDP-43 by RNAi (compare lanes 2 and 3 with lane 1, bottom 2 panels of Fig. 4c). Furthermore, the amount of TDP-43 protein, as examined by Western blotting, in the CYFIP1-pulled down RNPs decreased upon FMRP depletion by RNAi (compare lanes 1 and 2 of the top 2 panels, Fig. 4c), but the amount of FMRP in the CYFIP1-pulled down RNAs was not altered by depletion of TDP-43 (compare lanes 1 and 3 of top 2 panels, Fig. 4c). These data suggest that FMRP and TDP-43 together repress the translation of Rac1 mRNA at the initiation step, and that this co-repression is achieved through the binding of TDP-43 protein to the 3′UTR of Rac1 mRNA followed by recruitment of the FMRP-CYFIP1 complex that interacts with eIF4E to inhibit translation initiation of Rac1 mRNA (Fig. 9; [48]).
Regulation of local translation of Rac1 mRNA by TDP-43 and FMRP in the hippocampal neurons
We first analyzed Rac1 mRNA localization in DIV 6 and DIV 14 hippocampal neuron dendrites. Imaging analysis of Rac1 mRNA was carried out by FISH using Rac1 mRNA probe 1 (green, Fig. 5a). As shown, most of Rac1 mRNA molecules of DIV 6 primary mouse hippocampal neurons were confined to the soma [Fig. 5a(i)], while the amount of dendritic Rac1 mRNA in DIV 14 hippocampal neurons was ~8 times higher than that in DIV 6 neurons [bar diagram, Fig. 5a(i)]. FISH analysis further indicated that there was no significant change in the amount of Rac1 mRNA in DIV 14 dendrites upon depletion of either TDP-43 or FMRP [Fig. 5a(ii)]. Based on the above, we investigated the regulation of local translation of Rac1 mRNA by TDP-43 and FMRP in DIV 14 neurons.
Rac1-3′UTR-mediated translational co-repression by TDP-43 and FMRP in hippocampal neuron dendrites. a Imaging analysis of Rac1 mRNA by FISH in DIV 6 and DIV 14 hippocampal neurons. i Hippocampal neurons at different DIVs were hybridized with Rac1 probe 1 (green, see Supplementary materials and methods for details) and anti-TDP-43 (magenta). The histogram (bottom) compares the levels of Rac1 mRNA, presented as fluorescence intensity per 10 µm of dendrites of transfected neurons. Each set of data was collected from 15 to 18 transfected neurons from three independent experiments. Error bars represent SEM and significant difference is represented by ***p < 0.0001 (Student’s t test). Scale bar 5 µm. ii FISH and immunofluorescence staining analysis of DIV 14 hippocampal neurons transfected with Sc, TDP-si or FMRP-si RNA oligo using Rac1 probe 1(green, see Supplementary materials and methods for details), anti-TDP-43 (magenta), and anti-FMRP (red). The histogram compares the levels of Rac1 mRNA. For each set of samples, the data were collected from 10 to 14 transfected neurons from three independent experiments. Error bars represent SEM. Scale bar 1 µm. Note that we calculated the fluorescence intensity in all FISH experiments using Rac1 anti-sense probe 1 (see Supplementary materials and methods for details) in comparison to the control. Furthermore, specificity of the FISH experiments was validated by the 60–70 % decrease of the fluorescence intensity upon RNAi knock down of Rac1 mRNA (Fig S5). b Imaging analysis of DIV 14 hippocampal neurons co-transfected with different RNAi oligos and pmyr-dEGFP-3′UTR as described in Supplementary materials and methods. The transfected neurons were probed with anti-TDP-43 or anti-FMRP. The distribution patterns of myr-dEGFP proteins expressed in transfected neurons are exemplified by the confocal microscope images (top 3 panels). Immunofluorescence from the endogenous TDP-43 protein (blue) and FMRP protein (red) (middle 3 panels) and DIC images (bottom 3 panels) of the same neurons are also shown. Fluorescence intensities of myr-dEGFP protein at near and distal dendrites (0–150 and 150–300 µm, left histogram) and soma (right histogram) were measured and normalized to Sc oligo-transfected neurons. A total of 20–25 neurons from three independent experiments were characterized. Error bars represent SEM and significant difference is represented by ***p < 0.0001 (Student’s t test). Scale bar 5 µm. Note that blockade of the action potentials with tetrodotoxin (TTX) significantly decreased expression of myr-dEGFP in distal dendrites of TDP-si or FMRP-si transfected neurons (data not shown), supporting that myr-dEGFP fluorescence patterns reflect local translation of myr-dEGFP-3′UTR mRNA
Protein synthesis reporters were first constructed in which the myr-dEGFP coding sequence was attached to Rac1-3′UTR or to different 3′UTR mutants. Since the myristoylation peptide (myr) and the short half-life of destabilizing dEGFP protein impeded myr-dEGFP synthesized in somata from diffusing to distal dendrites, GFP fluorescence intensities in the distal dendritic regions would reflect the local protein synthesis [1]. As seen, in spite of there being no increase in Rac1 mRNA in DIV 14 dendrites upon depletion of TDP-43 and FMRP [Fig. 5a(ii)], a marked increase in translation was evident in the dendrites and somata (Fig. 5b), confirming the role of TDP-43 and FMRP in the inhibition of dendritic as well as somatic translation of Rac1 mRNA. Remarkably, when the localizations of Rac1 mRNA, TDP-43, FMRP and CYFIP1 in the dendrites of DIV 14 primary neurons were analyzed by combined FISH and immunofluorescence staining, the co-localization of CYFIP1 with Rac1 mRNA was significantly reduced upon either TDP-43 depletion or FMRP depletion (right set of histogram, Fig. 6). Notably, co-localization of FMRP with Rac1 mRNA was reduced upon depletion of TDP-43, but the reverse was not true (compare the left and middle sets of the histogram, Fig. 6). These data along with those shown in Figs. 2 and 3 indicate that recruitment of FMRP and CYFIP1 proteins to Rac1-3′UTR by TDP-43 is required for inhibition of localized dendritic translation of Rac1 mRNA in hippocampal neurons.
TDP-43 assisted recruitment of FMRP in dendritic Rac1 mRNP complex. Co-localization of Rac1 mRNA with TDP-43, FMRP and CYFIP1 proteins in TDP-43- or FMRP-depleted primary hippocampal neurons. DIV 14 hippocampal neurons were subjected to imaging analysis by FISH using of Rac1 mRNA using probe 1 (see Supplementary materials and methods for details) and immunofluorescence staining using anti-TDP-43, anti-FMRP or anti-CYFIP1. Representative confocal microscope images are shown on the left with arrows indicating co-localization of Rac1 mRNA with the three proteins. Quantification of the co-localization data from three sets of independent experiments (15–20 dendrites for each condition) is represented as a bar diagram on the right showing the percentages of dendritic Rac1 mRNA co-localized with TDP-43, FMRP or CYFIP1 proteins. Scale bar 5 μm. Significant differences are represented by ***p < 0.0001 and **p < 0.001 (Student’s t test)
Other mRNA targets of translational co-repression by TDP-43 and FMRP
FMRP was known to suppress the translation of several mRNAs important for synaptic plasticity including Rac1 mRNA, the impairment of which in part contributed to the development of Fragile X Syndrome (FXS) [6]. Therefore, we investigated whether, besides Rac1 mRNA, TDP-43 also played a role in regulating the translation of other FMRP targets including PSD-95 (Dlg4) mRNA, Map1b mRNA, GluR1 (Gria1) mRNA and CamKII mRNA [47, 72] in comparison to a non-target, i.e., mTOR mRNA. Of these targets, TDP-43 neither formed a complex with PSD-95 mRNA, nor had it any effect on the interaction of FMRP with this mRNA [Fig. 2a(ii)].
As shown by RNA IP of extracts from DIV 6 primary hippocampal neurons transfected with control Sc oligo, TDP-si oligo or FMRP-si oligo, mRNAs encoding Map1b, GluR1 and mTOR all interacted with TDP-43 and FMRP, whereas CamKII mRNA interacted only with FMRP [lanes 1 and 4, Fig. 7a(i)]. Moreover, effective recruitment of FMRP to Map1b mRNA, GluR1 mRNA or mTOR mRNA, but not CamKII mRNA, required TDP-43 [compare lanes 4 and 5, Fig. 7a(i)]. Notably, depletion of either TDP-43 or FMRP caused increased levels of Map1b and GluR1 proteins [Fig. 7a(ii)]. To examine the roles of TDP-43 and FMRP in the regulation of Map1b and GluR1 at the translational level, the distribution patterns of the different mRNAs in the sucrose gradients of cell extracts from primary hippocampal neurons transfected with the Sc, TDP-si or FMRP-si oligos were analyzed [Fig. 7b(i)]. In an interesting parallel to the Rac1 mRNA data [Fig. 1a(ii)], polysomal accumulation of Map1b mRNA and GluR1 mRNA at the expense of their association with 40S and 60S/80S monosomes was evident in TDP-43-depleted or FMRP-depleted neurons [see profiles representing results of three technical repeats from single biological sample in Fig. 7b(i) and histo bar diagram representing results from three biological samples in Fig. 7b(ii)], indicative of activation of translation upon removal of either FMRP or TDP-43. In contrast, polysomal accumulations of CamKII mRNA and PSD-95 mRNA were only evident in FMRP-depleted neurons (Fig. 7b), suggesting no role for TDP-43 in FMRP-mediated translational inhibition of these two mRNAs. Finally, neither TDP-43 nor FMRP had any role in the regulation of translation of mTOR mRNA (Fig. 7b).
Binding and translational regulation of dendritic mRNAs by FMRP and TDP-43. a i RNA-IP analysis of binding of GluR1(Gria1), Map1b, CamKII and mTOR mRNAs with FMRP and TDP-43 proteins in hippocampal neurons transfected with different RNAi oligos. Co-immunoprecipitated RNAs were analyzed by semi-quantitative (left) or quantitative (right) RT-PCR. Gapdh served as the negative control (not shown). Representative gel picture of semi-quantitative RT-PCR analysis are depicted on the left with lanes 1, 4 and 7 showing RNA enrichment in Sc oligo; lanes 2, 5 and 8 showing RNA enrichment in TDP-si oligo; and lanes 3, 6 and 9 showing RNA enrichment in FMRP-si oligo transfected neurons. Statistical analysis of data from three independent experiments is shown on the right. Significant differences in fold enrichment of different mRNAs between anti-IgG and anti-TDP-43 or anti-FMRP pulled down samples are represented by ***p < 0.0001 (Student’s t test). ii Relative expression levels of Map1b, GluR1, TDP-43 and FMRP proteins in cultured hippocampal neurons upon depletion of TDP-43 or FMRP by RNAi. Total proteins from DIV 6 primary hippocampal cultures transfected with different RNAi oligos for 48 h were subjected to Western blotting. Statistical analysis from three independent experiments is shown on the right. Significant differences are represented by ***p < 0.0001 (Student’s t test). b Polysomal distributions of different mRNAs in cultured DIV 6 hippocampal neurons transfected with different RNAi oligos. i RT-PCR analysis of RNAs isolated from different sucrose gradient fractions of cytoplasmic extracts from the transfected neurons. The data were plotted as the percentage of total mRNA in the gradient. Error bars represent SD from three experimental repeats from same set of biological sample. Percentage of mRNA level was significantly different between Sc oligo transfected and TDP-si or FMRP-si transfected neurons at 2nd–4th, 8th and 9th fractions for Map1b and GluR1 mRNA distributions. It was also significantly different between Sc oligo transfected and FMRP-si transfected neurons at 3rd, 4th and 9th fractions for CamKII and PSD-95 mRNA distributions (p < 0.01, pairwise t test). ii Quantitative RT-PCR analysis of the indicated mRNAs in polysome fractions of DIV 6 primary hippocampal neurons transfected with Sc, TDP-si and FMRP-si oligos, respectively. The fold of changes relative to the Sc oligo-transfected neurons are represented as the bar diagram. Error bars represent SD from three independent experiments with different biological samples. Significant differences between Sc oligo transfected and TDP-si or FMRP-si transfected neurons are represented by ***p < 0.0001 and **p < 0.001 (Student’s t test). Inputs are similar (data not shown)
Interestingly, as exemplified in Fig. 8a(i) and quantitatively analyzed in Fig. 8a(ii), the translational co-repression of Rac1, GluR1 and Map1b mRNAs by TDP-43 and FMRP was further supported by the rescue of exogenous GFP-FMRP-mediated decreases of Rac1, GluR1 and Map1b proteins, but not PSD-95 (data not shown), in DIV 14 primary hippocampal neurons upon depletion of TDP-43. Thus, of the five FMRP-regulated target mRNAs we analyzed, TDP-43 participated in translational repression of three of them, i.e., Rac1, Map1b, and GluR1, but not the other two, i.e., CamKII and PSD-95. Furthermore, as seen in Fig. 4c and Fig S6, Rac1 mRNA, GluR1 mRNA and Map1b mRNA were also associated with CYFIP1-containing RNP complexes in hippocampal neurons in a TDP-43-dependent and FMRP-dependent manner.
Co-operation of TDP-43 and FMRP in translational inhibition of different proteins and spinogenesis in primary hippocampal neurons. a Imaging analysis of the effect of TDP-43 depletion on FMRP-mediated inhibition of Rac1, Map1b and GluR1 protein expression in DIV 14 neurons. Representative pictures of hippocampal neurons co-transfected with pGFP or pGFP-FMRP plus RNA oligo Sc or TDP-si and showing the fluorescence patterns from GFP (green) and from immunofluorescence staining of TDP-43 (red), Rac1 (orange), GluR1 (blue) and Map1b (white), respectively, are displayed in (i). The DIC images of the neurons are shown on the right. Scale bars = 2 μm. ii The immunofluorescence intensities of Rac1, GluR1 and Map1b in 45–55 transfected neurons from three independent experiments were measured and normalized to those of the (Sc + GFP) neurons. The fold of changes were calculated and shown in the bar diagram. Error bars represent SEM. b Imaging analysis of the effect of TDP-43 depletion on FMRP-mediated reduction of the dendritic protrusion density. Representative confocal microscope images of DIV 14 hippocampal neurons transfected with Sc oligo + pGFP (top), Sc oligo + pGFP-FMRP (middle) and TDP-si oligo + pGFP-FMRP (bottom), respectively, are shown on the left, with arrows indicating the dendritic protrusions. Histoplot of the average densities of dendritic protrusion of 10–15 neurons from three independent experiments is shown on the right. Scale bars 5 μm. c Developmental changes in the expression levels of TDP-43, FMRP and Rac1 proteins in mouse primary hippocampal neurons in culture. Western blotting patterns are shown on the left and statistical analysis is shown on the right. Data represent the mean ± SD (error bars; N = 3). Significant differences in a, b and c were determined by one way ANOVA ***p < 0.0001, **p < 0.001 and *p < 0.01. d Histoplot showing the increased density of dendritic protrusions in cultured DIV 6 hippocampal neurons upon RNAi knockdown of TDP-43 or FMRP. For each set, 12–16 neurons from three independent experiments have been analyzed. Statistical significances are represented by **p < 0.001 and *p < 0.01 (Student’s t test)
Functionally, the translational co-repression by FMRP and TDP-43 regulated the spinogenesis of the hippocampal neurons. As shown in Fig. 8b, over-expression of GFP-FMRP decreased the spine density of primary hippocampal neurons, but this decrease could be rescued by depletion of TDP-43. Thus, the co-operation of TDP-43 with FMRP to repress the translation of a class of mRNAs in neurons at the initiation step via FMRP-bound CYFIP1 is indeed physiologically important with respect to the regulation of synaptic plasticity during brain development.
Discussion
Translational repression of dendritic mRNAs is an integral part of the regulatory program of neurons to ensure appropriate timing and place of the expression of specific dendritic proteins, thereby allowing proper spinogenesis and synapse formation for neurodevelopment [21, 43]. Impairment of this process, such as in FXS through silencing of the translational repressor FMRP, would lead to relief of translational repression of many mRNAs and development of neurological disorders [18, 59]. In this study, a functional and mechanistic synergy between FMRP and the major ALS/FTLD pathology related protein TDP-43 in the translational regulation of a subclass of their common mRNA targets important for spinogenesis, synaptic plasticity and/or neurodevelopment, i.e., Rac1, Map1b and GluR1, has been revealed. This synergy correlates well with previous findings of co-localization of FMRP and TDP-43 in RNPs in hippocampal neurons [67] and their capability to physically interact with each other [70]. Even more importantly, our data provide the molecular basis of a link between neurodevelopmental disorders and TDP-43 proteinopathies.
Initial evidence for the involvement of TDP-43 in translational repression is that in mouse primary hippocampal neurons (Fig. 1a) and a non-neuronal HEK293T cell line (Fig. 1b), TDP-43 inhibits the distribution of Rac1 mRNA in the polysomes and represses the translation of Rac1 mRNA [Fig S1c]. Furthermore, the regulation occurs through binding of TDP-43 to the UG/GU-rich sequences in the 3′UTR of Rac1 mRNA via its RRM1/RRM2 domain and subsequent recruitment of FMRP to Rac1 mRNP complex (Figs. 2, 3). More importantly, this regulation is dependent on FMRP [Fig. 1b(ii)], likely through physical interaction between FMRP and the glycine-rich domain of TDP-43 (Figs. 3c, S3a). Note that in the luciferase reporter assay of translation in HEK293T cells, over-expression of FMRP did not inhibit translation in the absence of exogenous expression of TDP-43 protein (Fig. 3c), likely due to the relatively very low level of endogenous TDP-43 in the HEK293T cells in comparison to mouse hippocampal neurons (Fig S3b).
FMRP represses translation through three different pathways [12, 55]. First, it can recruit specific miRNA complexes to inhibit translation of specific mRNAs, e.g., PSD-95, in neurons in an activity-dependent manner [51]. Secondly, FMRP can inhibit translation at the elongation stage by directly interacting with the translating ribosomes [17]. In this case FMRP protein was found to be mostly co-sedimented with higher polysome fractions [11, 60]. Finally, and mostly observed in synaptosomes and activated neurons, FMRP is primarily non-polysomic in nature, gets fractionated with translation initiation complex and mRNPs, and inhibits the initiation of translation [37, 72]. In the latter scenario, FMRP binds specific mRNAs such as Rac1, Map1b, CamKII, etc. [48], either directly to the G-quadruplex motif in 3′UTR or indirectly with the help of as yet to be unidentified factors [12]. This binding leads to the physical interaction of the FMRP-binding factor CYFIP1 with the initiation factor eIF4E at the 5′-cap site of the FMRP-bound mRNA. The formation of the eIF4E-CYFIP1-FMRP complex inhibits ribosome entry to the 5′-cap site of the mRNAs [48]. Interestingly, the rate-limiting step of translational co-repression of Rac1 mRNA by TDP-43 and FMRP in primary hippocampal neurons and HEK293T cells appears to be at the initiation stage (Fig. 4a, b) and it requires FMRP-TDP-43 interaction (Fig. 3c). Furthermore, this translational co-repression is closely associated with complex formation of CYFIP1-FMRP-TDP-43 (Fig. 4c) and between dendritic Rac1 mRNA with FMRP, TDP-43 and CYFIP1 in hippocampal neurons (Figs. 2, 4c and 6). Thus, our study provides the first evidence that repression of mRNAs lacking the G-quadruplex motif, such as Rac1 mRNA (Figs. 1, 3, 4, 5b and 8a) and GluR1 mRNA (Figs. 7, 8a), requires TDP-43, which acts as an adaptor to recruit the repression complex FMRP-CYFIP1 [see model of Fig. 9b]. The physiological importance of this co-regulatory scheme by the two RBPs is further exemplified by data shown in Fig. 8b–d.
A model of translational co-repression of specific mRNA(s) by FMRP and TDP-43. a Schematic representation of TDP-43 engaged in binding with mRNA(s) and FMRP protein. TDP-43 (red) binds to UG/GU motifs (black) in the 3′UTR or CDS of the mRNA(s) through its RNA-binding domains (RRM1, RRM2). RNA-bound TDP-43 also physically interacts with FMRP protein (yellow) through its glycine-rich domain and recruits FMRP to the vicinity of the mRNA(s). b ON–OFF states of the translation of specific mRNA(s) mediated by FMRP and TDP-43. In the “OFF” state, RNA-bound TDP-43 (red) recruits FMRP (yellow) or the FMRP-CYFIP1 complex to the vicinity of the mRNA(s). FMRP-bound CYFIP1 (green) interacts with eIF4E (blue), thereby blocking eIF4G (grey) from binding to eIF4E and forming the eIF4F translation initiation complex (not shown). Under FMRP-depleted conditions (left lower scheme), TDP-43 still binds to UG/GU repeat sequence(s) in the mRNA(s) but CYFIP1 (green) can no longer be recruited, thus allowing the formation of the eIF4E-eIF4G complex at the 5′cap site and ribosome entry to start translation. When TDP-43 is depleted (right lower scheme) the inhibitory complex CYFIP1-FMRP cannot be recruited to the vicinity of the mRNA(s) to repress translation. Note that in either of the “ON” states, initiation complex formation between eIF4E and eIF4G can still be inhibited by the drug 4EGI-1
Why would FMRP need an adaptor protein like TDP-43 in the physiological system to select specific mRNA targets for CYFIP1-mediated translational repression when it has its own capacity to bind mRNAs? There are several advantageous possibilities. First, FMRP is not likely to recognize and bind all mRNAs effectively [12, 72]. TDP-43 could help recruit the FMRP-CYFIP1 complex to the vicinities of mRNAs whether they contain the G-quadruplex motif (e.g., Map1b, Figs. 7a and S6a) or not (e.g., Rac1 and GluR1, Figs. 2, 3a, 4c, 6, 7a and S6). Second, the requirement for two different RNA-binding proteins in translational co-repression allows fine-tuning of the expression of a class of mRNAs at the translational level, thereby facilitating regulation of normal development. For instance, the level of FMRP in hippocampal neurons remains relatively unchanged or increases gradually during later stages of embryonic development [35], as well as during in vitro differentiation (Fig. 8c). This could be required for maintenance of the overall expression profiles of FMRP target genes. In contrast, the level of TDP-43 gradually decreases, while that of Rac1, a positive regulator of spinogenesis [13] increases as primary neurons mature in culture (Fig. 8c; [45]). Decreased TDP-43 would mitigate TDP-43-FMRP-CYFIP1-eIF4E-mediated translational repression of dendritic Rac1 mRNA (see model in Fig. 9b), thus facilitating spinogenesis during neuron maturation (Fig. 8b, d) and brain development [13, 45].
The functional synergy of FMRP and TDP-43 in translational regulation through their physical association in specific mRNPs (Fig. 9) provides a molecular link between a range of neurodevelopmental disorders and several neurodegenerative diseases associated with TDP-43 proteinopathies. In particular, a few neurodevelopmental disorders exhibit pathological phenotypes overlapping with those of FXS, but without apparent involvement of FMRP. For example, genetically and biologically, ~20 % of gene/RNA targets of FMRP overlap with genes associated with ASDs, whereas only up to 5 % of ASD patients carry any type of mutation at the FMR1 gene [2, 19]. Notably, neurodegeneration has been reported for a number of ASD cases [32] while the molecular basis is unclear. Moreover, based on genetic linkage and functional properties, CYFIP1/2 proteins are good causal candidates for autism, though a precise functional role remains to be identified [3]. These studies together indicate the involvement of unknown functionally interacting partner(s) of FMRP and CYFIP in ASD cases lacking a known gene mutation. Our data suggest that TDP-43 might be one of the candidate partners. Based on the literature (Table S1), we also speculate that loss-of-function of either TDP-43 or FMRP leading to the relief of translational repression of a subset of their common target mRNAs may underlie the pathogenesis of a subclass of AD and Schizophrenia. In view of the apparently essential role of FMRP in neuronal functions suggested by studies of acute knock down of FMRP in mice [26] or in cell cultures ([23], current study), the phenotypes of Fmr1 KO mice [31] should be more severe than currently known. Perhaps the congenital absence of Fmr1 in these Fmr1 KO mice and in FXS patients as well has induced some compensatory programs or factors in vivo [36]. It would be interesting to carry out a comparative study of hippocampal neurons from the Fmr1 KO mice, to see whether the co-operative scenario of FMRP and TDP-43 in translational co-repression, as depicted in Fig. 9, is replaced by other regulatory scenarios in these neurons. Finally, several ALS-associated mutations across the glycine-rich domain of TDP-43 alter its interaction with FMRP (data not shown). Whether impairment of the FMRP-TDP-43 interaction has any role in the variance in gene expression related to ALS pathology could also be an interesting avenue of future research.
We endeavored to identify putative common mRNA targets of TDP-43 and FMRP, other than those already identified, some of which might be co-regulated translationally by these two RNA binding factors. A comparative analysis of the databases of TDP-43-bound mRNAs and FMRP-bound mRNAs (Table S2) has identified 1140 mRNAs as possible common targets of TDP-43 and FMRP. Gene ontology analysis identified 160 of these 1140 mRNAs as being important for neuron structure, function and development (Table S3). However, we could not determine, at this moment, which of these 160 mRNAs are co-regulated by TDP-43 and FMRP at the translational level, except for Rac1 mRNA, GluR1 mRNA and Map1b mRNA (as reported above). Interestingly, this gene list also includes candidate genes for ASD (e.g., Reln, Shank3, Rac1, Mapk1 etc.), for AD (e.g., App, Apoe, Ank1, etc.), and for schizophrenia (e.g., Gsk3b, Grin2b, etc.). This bioinformatics analysis further strengthens the notion that FMRP and TDP-43 collaboratively contribute to the pathogenesis of different neurodevelopmental disorders and neurodegenerative diseases by co-regulating the translation of certain members of a common set of their target mRNAs.
In summary, our study has uncovered a paradigm scenario of how TDP-43 regulates the translation of specific mRNAs. Furthermore, for the first time, we reveal a functional synergy between FMRP and TDP-43 in translational co-repression of a class of neuronal genes important for the regulation of spinogenesis, neuron architecture and neuronal plasticity. Loss-of-function of this synergy is likely to play a significant role in the pathogenesis of FMRP-associated neurological disorders, such as ASD and AD, and TDP-43-associated neurodegenerative diseases. Since mutations or mis-expression of either factor would affect a common set of target genes, it will be important to examine whether mutations or mis-metabolism of TDP-43 exist in a subset of ASD, AD or schizophrenia cases.
Abbreviations
- TDP-43:
-
TAR DNA binding protein, 43 kDa
- FMRP:
-
Fragile X mental retardation protein
- CYFIP1:
-
Cytoplasmic FMRP interacting protein 1
- FISH:
-
Fluorescence in situ hybridization
- RNA-IP:
-
RNA immunoprecipitation
- DIV:
-
Day in vitro
- DIC:
-
Differential interference contrast
References
Aakalu G, Smith WB, Nguyen N, Jiang C, Schuman EM (2001) Dynamic visualization of local protein synthesis in hippocampal neurons. Neuron 30:489–502
Abbeduto L, McDuffie A, Thurman AJ (2014) The fragile X syndrome-autism comorbidity: what do we really know? Front Genet 5:355. doi:10.3389/fgene.2014.00355
Abekhoukh S, Bardoni B (2014) CYFIP family proteins between autism and intellectual disability: links with Fragile X syndrome. Front Cell Neurosci 8:81. doi:10.3389/fncel.2014.00081
Baralle M, Buratti E, Baralle FE (2013) The role of TDP-43 in the pathogenesis of ALS and FTLD. Biochem Soc Trans 41:1536–1540. doi:10.1042/BST20130186
Bechara EG, Didiot MC, Melko M, Davidovic L, Bensaid M, Martin P, Castets M, Pognonec P, Khandjian EW, Moine H et al (2009) A novel function for fragile X mental retardation protein in translational activation. PLoS Biol 7:e16. doi:10.1371/journal.pbio.1000016
Bongmba OY, Martinez LA, Elhardt ME, Butler K, Tejada-Simon MV (2011) Modulation of dendritic spines and synaptic function by Rac1: a possible link to Fragile X syndrome pathology. Brain Res 1399:79–95. doi:10.1016/j.brainres.2011.05.020
Bose JK, Wang IF, Hung L, Tarn WY, Shen CK (2008) TDP-43 overexpression enhances exon 7 inclusion during the survival of motor neuron pre-mRNA splicing. J Biol Chem 283:28852–28859. doi:10.1074/jbc.M805376200
Bray N (2016) Neurodegenerative disease: repeating mistakes. Nat Rev Neurosci. doi:10.1038/nrn.2016.64
Brown V, Jin P, Ceman S, Darnell JC, O’Donnell WT, Tenenbaum SA, Jin X, Feng Y, Wilkinson KD, Keene JD et al (2001) Microarray identification of FMRP-associated brain mRNAs and altered mRNA translational profiles in fragile X syndrome. Cell 107:477–487
Buffington SA, Huang W, Costa-Mattioli M (2014) Translational control in synaptic plasticity and cognitive dysfunction. Annu Rev Neurosci 37:17–38. doi:10.1146/annurev-neuro-071013-014100
Ceman S, O’Donnell WT, Reed M, Patton S, Pohl J, Warren ST (2003) Phosphorylation influences the translation state of FMRP-associated polyribosomes. Hum Mol Genet 12:3295–3305. doi:10.1093/hmg/ddg350
Chen E, Joseph S (2015) Fragile X mental retardation protein: a paradigm for translational control by RNA-binding proteins. Biochimie 114:147–154. doi:10.1016/j.biochi.2015.02.005
Corbetta S, Gualdoni S, Ciceri G, Monari M, Zuccaro E, Tybulewicz VL, de Curtis I (2009) Essential role of Rac1 and Rac3 GTPases in neuronal development. FASEB J 23:1347–1357. doi:10.1096/fj.08-121574
Coyne AN, Siddegowda BB, Estes PS, Johannesmeyer J, Kovalik T, Daniel SG, Pearson A, Bowser R, Zarnescu DC (2014) Futsch/MAP1B mRNA is a translational target of TDP-43 and is neuroprotective in a Drosophila model of amyotrophic lateral sclerosis. J Neurosci 34:15962–15974. doi:10.1523/JNEUROSCI.2526-14.2014
Coyne AN, Yamada SB, Siddegowda BB, Estes PS, Zaepfel BL, Johannesmeyer JS, Lockwood DB, Pham LT, Hart MP, Cassel JA et al (2015) Fragile X protein mitigates TDP-43 toxicity by remodeling RNA granules and restoring translation. Hum Mol Genet 24:6886–6898. doi:10.1093/hmg/ddv389
Darnell JC, Klann E (2013) The translation of translational control by FMRP: therapeutic targets for FXS. Nat Neurosci 16:1530–1536. doi:10.1038/nn.3379
Darnell JC, Van Driesche SJ, Zhang C, Hung KY, Mele A, Fraser CE, Stone EF, Chen C, Fak JJ, Chi SW et al (2011) FMRP stalls ribosomal translocation on mRNAs linked to synaptic function and autism. Cell 146:247–261. doi:10.1016/j.cell.2011.06.013
De Rubeis S, Bagni C (2011) Regulation of molecular pathways in the Fragile X Syndrome: insights into Autism Spectrum Disorders. J Neurodev Disord 3:257–269. doi:10.1007/s11689-011-9087-2
Ebert DH, Greenberg ME (2013) Activity-dependent neuronal signalling and autism spectrum disorder. Nature 493:327–337. doi:10.1038/nature11860
El Fatimy R, Tremblay S, Dury AY, Solomon S, De Koninck P, Schrader JW, Khandjian EW (2012) Fragile X mental retardation protein interacts with the RNA-binding protein caprin1 in neuronal RIBONUCLEOPROTEIN complexes [corrected]. PLoS One 7:e39338. doi:10.1371/journal.pone.0039338
Fernandez-Moya SM, Bauer KE, Kiebler MA (2014) Meet the players: local translation at the synapse. Front Mol Neurosci 7:84. doi:10.3389/fnmol.2014.00084
Fernandez E, Rajan N, Bagni C (2013) The FMRP regulon: from targets to disease convergence. Front Neurosci 7:191. doi:10.3389/fnins.2013.00191
Ferron L, Nieto-Rostro M, Cassidy JS, Dolphin AC (2014) Fragile X mental retardation protein controls synaptic vesicle exocytosis by modulating N-type calcium channel density. Nat Commun 5:3628. doi:10.1038/ncomms4628
Freibaum BD, Chitta RK, High AA, Taylor JP (2010) Global analysis of TDP-43 interacting proteins reveals strong association with RNA splicing and translation machinery. J Proteome Res 9:1104–1120. doi:10.1021/pr901076y
Gerstberger S, Hafner M, Tuschl T (2014) A census of human RNA-binding proteins. Nat Rev Genet 15:829–845. doi:10.1038/nrg3813
Guo W, Polich ED, Su J, Gao Y, Christopher DM, Allan AM, Wang M, Wang F, Wang G, Zhao X (2015) Fragile X proteins FMRP and FXR2P control synaptic GluA1 expression and neuronal maturation via distinct mechanisms. Cell Rep 11:1651–1666. doi:10.1016/j.celrep.2015.05.013
Ishizuka A, Siomi MC, Siomi H (2002) A Drosophila fragile X protein interacts with components of RNAi and ribosomal proteins. Genes Dev 16:2497–2508. doi:10.1101/gad.1022002
Jayaseelan S, Tenenbaum SA (2012) Neurodevelopmental disorders: signalling pathways of fragile X syndrome. Nature 492:359–360. doi:10.1038/nature11764
Jovicic A, Paul JW 3rd, Gitler AD (2016) Nuclear transport dysfunction: a common theme in amyotrophic lateral sclerosis and frontotemporal dementia. J Neurochem. doi:10.1111/jnc.13642
Kapeli K, Yeo GW (2012) Genome-wide approaches to dissect the roles of RNA binding proteins in translational control: implications for neurological diseases. Front Neurosci 6:144. doi:10.3389/fnins.2012.00144
Kazdoba TM, Leach PT, Silverman JL, Crawley JN (2014) Modeling fragile X syndrome in the Fmr1 knockout mouse. Intractable Rare Dis Res 3:118–133. doi:10.5582/irdr.2014.01024
Kern JK, Geier DA, Sykes LK, Geier MR (2013) Evidence of neurodegeneration in autism spectrum disorder. Transl Neurodegener 2:17. doi:10.1186/2047-9158-2-17
Khandjian EW, Huot ME, Tremblay S, Davidovic L, Mazroui R, Bardoni B (2004) Biochemical evidence for the association of fragile X mental retardation protein with brain polyribosomal ribonucleoparticles. Proc Natl Acad Sci 101:13357–13362. doi:10.1073/pnas.0405398101
Krichevsky AM, Kosik KS (2001) Neuronal RNA granules: a link between RNA localization and stimulation-dependent translation. Neuron 32:683–696
Kroon T, Sierksma MC, Meredith RM (2013) Investigating mechanisms underlying neurodevelopmental phenotypes of autistic and intellectual disability disorders: a perspective. Front Syst Neurosci 7:75. doi:10.3389/fnsys.2013.00075
La Fata G, Gartner A, Dominguez-Iturza N, Dresselaers T, Dawitz J, Poorthuis RB, Averna M, Himmelreich U, Meredith RM, Achsel T et al (2014) FMRP regulates multipolar to bipolar transition affecting neuronal migration and cortical circuitry. Nat Neurosci 17:1693–1700. doi:10.1038/nn.3870
Laggerbauer B, Ostareck D, Keidel EM, Ostareck-Lederer A, Fischer U (2001) Evidence that fragile X mental retardation protein is a negative regulator of translation. Hum Mol Genet 10:329–338
Lagier-Tourenne C, Polymenidou M, Cleveland DW (2010) TDP-43 and FUS/TLS: emerging roles in RNA processing and neurodegeneration. Hum Mol Genet 19:R46–R64. doi:10.1093/hmg/ddq137
Lai MC, Lee YH, Tarn WY (2008) The DEAD-box RNA helicase DDX3 associates with export messenger ribonucleoproteins as well as tip-associated protein and participates in translational control. Mol Biol Cell 19:3847–3858. doi:10.1091/mbc.E07-12-1264
Lee A, Li W, Xu K, Bogert BA, Su K, Gao FB (2003) Control of dendritic development by the Drosophila fragile X-related gene involves the small GTPase Rac1. Development 130:5543–5552. doi:10.1242/dev.00792
Lee EB, Lee VM, Trojanowski JQ (2012) Gains or losses: molecular mechanisms of TDP43-mediated neurodegeneration. Nat Rev Neurosci 13:38–50. doi:10.1038/nrn3121
Ling SC, Polymenidou M, Cleveland DW (2013) Converging mechanisms in ALS and FTD: disrupted RNA and protein homeostasis. Neuron 79:416–438. doi:10.1016/j.neuron.2013.07.033
Liu-Yesucevitz L, Bassell GJ, Gitler AD, Hart AC, Klann E, Richter JD, Warren ST, Wolozin B (2011) Local RNA translation at the synapse and in disease. J Neurosci 31:16086–16093. doi:10.1523/JNEUROSCI.4105-11.2011
Maiti P, Manna J, Ilavazhagan G, Rossignol J, Dunbar GL (2015) Molecular regulation of dendritic spine dynamics and their potential impact on synaptic plasticity and neurological diseases. Neurosci Biobehav Rev 59:208–237. doi:10.1016/j.neubiorev.2015.09.020
Majumder P, Chen YT, Bose JK, Wu CC, Cheng WC, Cheng SJ, Fang YH, Chen YL, Tsai KJ, Lien CC et al (2012) TDP-43 regulates the mammalian spinogenesis through translational repression of Rac1. Acta Neuropathol 124:231–245. doi:10.1007/s00401-012-1006-4
Menon L, Mader SA, Mihailescu MR (2008) Fragile X mental retardation protein interactions with the microtubule associated protein 1B RNA. RNA 14:1644–1655. doi:10.1261/rna.1100708
Muddashetty RS, Kelic S, Gross C, Xu M, Bassell GJ (2007) Dysregulated metabotropic glutamate receptor-dependent translation of AMPA receptor and postsynaptic density-95 mRNAs at synapses in a mouse model of fragile X syndrome. J Neurosci 27:5338–5348. doi:10.1523/JNEUROSCI.0937-07.2007
Napoli I, Mercaldo V, Boyl PP, Eleuteri B, Zalfa F, De Rubeis S, Di Marino D, Mohr E, Massimi M, Falconi M et al (2008) The fragile X syndrome protein represses activity-dependent translation through CYFIP1, a new 4E-BP. Cell 134:1042–1054. doi:10.1016/j.cell.2008.07.031
Neumann M, Sampathu DM, Kwong LK, Truax AC, Micsenyi MC, Chou TT, Bruce J, Schuck T, Grossman M, Clark CM et al (2006) Ubiquitinated TDP-43 in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Science 314:130–133. doi:10.1126/science.1134108
Osterweil EK, Krueger DD, Reinhold K, Bear MF (2010) Hypersensitivity to mGluR5 and ERK1/2 leads to excessive protein synthesis in the hippocampus of a mouse model of fragile X syndrome. J Neurosci 30:15616–15627. doi:10.1523/JNEUROSCI.3888-10.2010
Plante I, Davidovic L, Ouellet DL, Gobeil LA, Tremblay S, Khandjian EW, Provost P (2006) Dicer-derived microRNAs are utilized by the fragile X mental retardation protein for assembly on target RNAs. J Biomed Biotechnol 2006:1–12. doi:10.1155/JBB/2006/64347
Polymenidou M, Lagier-Tourenne C, Hutt KR, Huelga SC, Moran J, Liang TY, Ling SC, Sun E, Wancewicz E, Mazur C et al (2011) Long pre-mRNA depletion and RNA missplicing contribute to neuronal vulnerability from loss of TDP-43. Nat Neurosci 14:459–468. doi:10.1038/nn.2779
Pozueta J, Lefort R, Ribe EM, Troy CM, Arancio O, Shelanski M (2013) Caspase-2 is required for dendritic spine and behavioural alterations in J20 APP transgenic mice. Nat Commun 4:1939. doi:10.1038/ncomms2927
Re A, Joshi T, Kulberkyte E, Morris Q, Workman CT (2014) RNA-protein interactions: an overview. Methods Mol Biol 1097:491–521. doi:10.1007/978-1-62703-709-9_23
Santoro MR, Bray SM, Warren ST (2012) Molecular mechanisms of fragile X syndrome: a twenty-year perspective. Annu Rev Pathol 7:219–245. doi:10.1146/annurev-pathol-011811-132457
Sekiyama N, Arthanari H, Papadopoulos E, Rodriguez-Mias RA, Wagner G, Leger-Abraham M (2015) Molecular mechanism of the dual activity of 4EGI-1: dissociating eIF4G from eIF4E but stabilizing the binding of unphosphorylated 4E-BP1. Proc Natl Acad Sci 112:E4036–E4045. doi:10.1073/pnas.1512118112
Sephton CF, Cenik C, Kucukural A, Dammer EB, Cenik B, Han Y, Dewey CM, Roth FP, Herz J, Peng J et al (2011) Identification of neuronal RNA targets of TDP-43-containing ribonucleoprotein complexes. J Biol Chem 286:1204–1215. doi:10.1074/jbc.M110.190884
Sephton CF, Yu G (2015) The function of RNA-binding proteins at the synapse: implications for neurodegeneration. Cell Mol Life Sci 72:3621–3635. doi:10.1007/s00018-015-1943-x
Sidorov MS, Auerbach BD, Bear MF (2013) Fragile X mental retardation protein and synaptic plasticity. Mol Brain 6:15. doi:10.1186/1756-6606-6-15
Stefani G, Fraser CE, Darnell JC, Darnell RB (2004) Fragile X mental retardation protein is associated with translating polyribosomes in neuronal cells. J Neurosci 24:7272–7276. doi:10.1523/JNEUROSCI.2306-04.2004
Tollervey JR, Curk T, Rogelj B, Briese M, Cereda M, Kayikci M, Konig J, Hortobagyi T, Nishimura AL, Zupunski V et al (2011) Characterizing the RNA targets and position-dependent splicing regulation by TDP-43. Nat Neurosci 14:452–458. doi:10.1038/nn.2778
Triller A, Sheng M (2012) Synaptic structure and function. Curr Opin Neurobiol 22:363–365. doi:10.1016/j.conb.2012.04.002
Vanden Broeck L, Callaerts P, Dermaut B (2014) TDP-43-mediated neurodegeneration: towards a loss-of-function hypothesis? Trends Mol Med 20:66–71. doi:10.1016/j.molmed.2013.11.003
Vasilyev N, Polonskaia A, Darnell JC, Darnell RB, Patel DJ, Serganov A (2015) Crystal structure reveals specific recognition of a G-quadruplex RNA by a beta-turn in the RGG motif of FMRP. Proc Natl Acad Sci 112:E5391–E5400. doi:10.1073/pnas.1515737112
Wang H (2015) Fragile X mental retardation protein: from autism to neurodegenerative disease. Front Cell Neurosci 9:43. doi:10.3389/fncel.2015.00043
Wang IF, Wu LS, Chang HY, Shen CK (2008) TDP-43, the signature protein of FTLD-U, is a neuronal activity-responsive factor. J Neurochem 105:797–806. doi:10.1111/j.1471-4159.2007.05190.x
Wang IF, Wu LS, Shen CK (2008) TDP-43: an emerging new player in neurodegenerative diseases. Trends Mol Med 14:479–485. doi:10.1016/j.molmed.2008.09.001
Wu LS, Cheng WC, Hou SC, Yan YT, Jiang ST, Shen CK (2010) TDP-43, a neuro-pathosignature factor, is essential for early mouse embryogenesis. Genesis 48:56–62. doi:10.1002/dvg.20584
Wu LS, Cheng WC, Shen CK (2012) Targeted depletion of TDP-43 expression in the spinal cord motor neurons leads to the development of amyotrophic lateral sclerosis-like phenotypes in mice. J Biol Chem 287:27335–27344. doi:10.1074/jbc.M112.359000
Yu Z, Fan D, Gui B, Shi L, Xuan C, Shan L, Wang Q, Shang Y, Wang Y (2012) Neurodegeneration-associated TDP-43 interacts with fragile X mental retardation protein (FMRP)/Staufen (STAU1) and regulates SIRT1 expression in neuronal cells. J Biol Chem 287:22560–22572. doi:10.1074/jbc.M112.357582
Zalfa F, Achsel T, Bagni C (2006) mRNPs, polysomes or granules: FMRP in neuronal protein synthesis. Curr Opin Neurobiol 16:265–269. doi:10.1016/j.conb.2006.05.010
Zalfa F, Giorgi M, Primerano B, Moro A, Di Penta A, Reis S, Oostra B, Bagni C (2003) The fragile X syndrome protein FMRP associates with BC1 RNA and regulates the translation of specific mRNAs at synapses. Cell 112:317–327
Acknowledgments
We thank Drs. Woan-Yuh Tarn and Yi-Shuian Huang (IBMS, Academia Sinica) for their generous gifts of experimental materials and advices. The expertise of Sue-Ping Lee and Shu-Mei Huang in the Microscopy Core at IMB as well as Dr. Po-Yen Lin in the Imaging Core facility of ICOB and all members of Bioinformatics Core at IMB are greatly appreciated. We sincerely convey our gratitude to Dr. John O’Brien from English Editing Service, IMB, Academia Sinica and our fellow lab mate Dr. Lien-Szu Wu for their helps in editing of the manuscript. This work was supported by the Frontier of Science Award from the National Science Council and a Senior Investigator Award from the Academia Sinica, Taipei, Taiwan (R.O.C.).
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made.
The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder.
To view a copy of this licence, visit https://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Majumder, P., Chu, JF., Chatterjee, B. et al. Co-regulation of mRNA translation by TDP-43 and Fragile X Syndrome protein FMRP. Acta Neuropathol 132, 721–738 (2016). https://doi.org/10.1007/s00401-016-1603-8
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00401-016-1603-8