Abstract
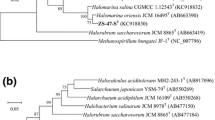
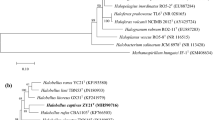
Halophilic archaeal strain R28T was isolated from the brown alga Laminaria produced at Dalian, Liaoning Province, China. The cells of the strain were pleomorphic and lysed in distilled water, stained Gram-negative, and formed red-pigmented colonies. Strain R28T was able to grow at 25–50 °C (optimum 42 °C), in the presence of 3.1–5.1 M NaCl (optimum 3.9 M NaCl), with 0.005–1.0 M MgCl2 (optimum 0.01 M MgCl2) and at pH 6.0–9.5 (optimum pH 7.0–7.5). The minimal NaCl concentration to prevent cell lysis was 15 % (w/v). The major polar lipids of the strain were identified as phosphatidic acid, phosphatidylglycerol, phosphatidylglycerol phosphate methyl ester, and two glycolipids chromatographically identical to those of Halovenus aranensis CGMCC 1.11001T. The 16S rRNA gene and rpoB′ gene of strain R28T were phylogenetically related to the corresponding genes of Hvn. aranensis CGMCC 1.11001T (91.9–97.2 and 82.9 % nucleotide identity, respectively). The DNA G+C content of strain R28T was determined to be 56.3 mol%. The phenotypic, chemotaxonomic, and phylogenetic properties suggest that strain R28T (=CGMCC 1.10592T = JCM 17269T) represents a novel species of the genus Halovenus, for which the name Halovenus rubra sp. nov. is proposed.

Similar content being viewed by others
References
Amoozegar MA, Makhdoumi-Kakhki A, Mehrshad M, Shahzadeh Fazeli SA, Ventosa A (2013) Halopenitus malekzadehii sp. nov., a novel extremely halophilic archaeon from a salt lake. Int J Syst Evol Microbiol 63:3232–3236
Collins MD (1985) Isoprenoid quinone analysis in bacterial classification and identification. In: Goodfellow M, Minnikin DE (eds) Chemical methods in bacterial systematics. Academic Press, London, pp 267–287
Cui H-L, Lin Z-Y, Dong Y, Zhou P-J, Liu S-J (2007) Halorubrum litoreum sp. nov., an extremely halophilic archaeon from a solar saltern. Int J Syst Evol Microbiol 57:2204–2206
Cui H-L, Zhou P-J, Oren A, Liu S-J (2009) Intraspecific polymorphism of 16S rRNA genes in two halophilic archaeal genera, Haloarcula and Halomicrobium. Extremophiles 13:31–37
Cui H-L, Gao X, Yang X, Xu X-W (2010) Halorussus rarus gen. nov., sp. nov., a new member of the family Halobacteriaceae isolated from a marine solar saltern. Extremophiles 14:493–499
Cui H-L, Yang X, Mou Y-Z (2011) Salinarchaeum laminariae gen. nov., sp. nov.: a new member of the family Halobacteriaceae isolated from salted brown alga Laminaria. Extremophiles 15:625–631
Dussault HP (1955) An improved technique for staining red halophilic bacteria. J Bacteriol 70:484–485
Echigo A, Minegishi H, Shimane Y, Kamekura M, Itoh T, Usami R (2013) Halomicroarcula pellucida gen. nov., sp. nov., a non-pigmented, transparent-colony-forming, halophilic archaeon isolated from solar salt. Int J Syst Evol Microbiol 63:3556–3562
Gonzalez C, Gutierrez C, Ramirez C (1978) Halobacterium vallismortis sp. nov. an amylolytic and carbohydrate-metabolizing, extremely halophilic bacterium. Can J Microbiol 24:710–715
Gutiérrez C, González C (1972) Method for simultaneous detection of proteinase and esterase activities in extremely halophilic bacteria. Appl Microbiol 24:516–517
Kates M, Kushwaha SC (1995) Isoprenoids and polar lipids of extreme halophiles. In: DasSarma S, Fleischmann EM (eds) Archaea, a laboratory manual. Halophiles. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, pp 35–54
McDade JJ, Weaver RH (1959) Rapid methods for the detection of gelatin hydrolysis. J Bacteriol 77:60–64
Makhdoumi-Kakhki A, Amoozegar MA, Ventosa A (2012) Halovenus aranensis gen. nov., sp. nov., an extremely halophilic archaeon from Aran-Bidgol salt lake. Int J Syst Evol Microbiol 62:1331–1336
Marmur J, Doty P (1962) Determination of the base composition of deoxyribonucleic acid from its thermal denaturation temperature. J Mol Biol 5:109–118
Minegishi H, Kamekura M, Itoh T, Echigo A, Usami R, Hashimoto T (2010) Further refinement of Halobacteriaceae phylogeny based on the full-length RNA polymerase subunit B′ (rpoB′) gene. Int J Syst Evol Microbiol 60:2398–2408
Oren A, Ventosa A, Grant WD (1997) Proposed minimal standards for description of new taxa in the order Halobacteriales. Int J Syst Bacteriol 47:233–238
Oren A, Arahal DR, Ventosa A (2009) Emended descriptions of genera of the family Halobacteriaceae. Int J Syst Evol Microbiol 59:637–642
Oren A (2012) Taxonomy of the family Halobacteriaceae: a paradigm for changing concepts in prokaryote systematics. Int J Syst Evol Microbiol 62:263–271
Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S (2011) MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28:2731–2739
Yim KJ, Cha I-T, Lee H-W, Song HS, Kim K-N, Lee S-J, Nam Y-D, Hyun D-W, Bae J-W, Rhee S-K, Seo M-J, Choi J-S, Choi H-J, Roh SW, Kim D (2014) Halorubrum halophilum sp. nov., an extremely halophilic archaeon isolated from a salt-fermented seafood. Antonie Van Leeuwenhoek 105:603–612
Acknowledgments
This work was supported by the National Natural Science Foundation of China (No. 31370054), the Grant from China Ocean Mineral Resources Research and Development Association (COMRA) Special Foundation (DY125-15-R-03), the Qinglan Project of Jiangsu Province and a project funded by the Priority Academic Program Development of Jiangsu Higher Education Institutions (PAPD).
Author information
Authors and Affiliations
Corresponding authors
Additional information
The GenBank/EMBL/DDBJ accession numbers for the 16S rRNA gene and rpoB ′ gene sequences of strain R28T are HM159605 and KF573409, respectively.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Han, D., Zhang, WJ., Cui, HL. et al. Halovenus rubra sp. nov., Isolated from Salted Brown Alga Laminaria . Curr Microbiol 70, 91–95 (2015). https://doi.org/10.1007/s00284-014-0685-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00284-014-0685-6