Abstract
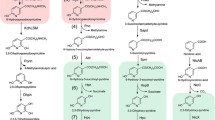
Pseudomonas putida J5 is an efficient nicotine-degrading bacterial strain isolated from the tobacco rhizosphere. We successfully performed a comprehensive whole-genome analysis of nicotine metabolism-associated genes by Tn5 transposon mutagenesis in P. putida J5. A total of 18 mutants with unique insertions screened from 16,324 Tn5-transformants failed to use nicotine as the sole carbon source. Flanking sequences of the Tn5 transposon were cloned with a shotgun method from all of the nicotine-growth-deficient mutants. The potentially essential products of mutated gene were classified as follows: oxidoreductases, protein and metal transport systems, proteases and peptidases, transcriptional and translational regulators, and unknown proteins. Bioinformatic analysis of the Tn5 insertion sites indicated that the nicotine metabolic genes were separated and widely distributed in the genome. One of the mutants, M2022, was a Tn5 insert into a gene encoding a homolog of 6-hydroxy-l-nicotine oxidase, the second enzyme of nicotine metabolism in Arthrobacter nicotinovorans. Genetic and biochemical analysis confirmed that three open reading frames (ORFs) from an approximately 13-kb fragment recovered from the mutant M2022 were responsible for the transformation of nicotine to 3-succinoyl-pyridine via pseudooxynicotine and 3-succinoyl semialdehyde-pyridine, the first three steps of nicotine degradation. Further research on these mutants and the Tn5-inserted genes will help us characterize nicotine metabolic processes in P. putida J5.




Similar content being viewed by others
References
Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ (1990) Basic local alignment search tool. J Mol Biol 215:403–410
Armstrong DW, Wang X, Ercal N (1998) Enantiomeric composition of nicotine in smokeless tobacco, medicinal products, and commercial reagents. Chiriality 10:587–591
Aubert D, Naas T, Héritier C, Poirel L, Nordmann P (2006) Functional characterization of IS1999, an IS4 family element involved in mobilization and expression of β-lactam resistance genes. J Bacteriol 188:6506–6514
Baitsch D, Sandu C, Brandsch R, Igloi GL (2001) Gene cluster on pAO1 of Arthrobacter nicotinovorans involved in degradation of the plant alkaloid nicotine: cloning, purification, and characterization of 2, 6-dihydroxypyridine 3-hydroxylase. J Bacteriol 183:5262–5267
Berg DE, Berg CM (1983) The prokaryotic transposable element Tn5. Nat Biotechnol 1:417–435
Brandsch R (2006) Microbiology and biochemistry of nicotine degradation. Appl Microbiol Biotechnol 69:493–498
Cánovas D, Cases I, de Lorenzo V (2003) Heavy metal tolerance and metal homeostasis in Pseudomonas putida as revealed by complete genome analysis. Environ Microbiol 5:1242–1256
Chandler M, Mahillon J (2002) Insertion sequences revisited. In: Craig NL, Craigie R, Geller M, Lambowitz AM (eds) Mobile DNA II. ASM Press, Washington D.C., pp 305–366
Chiribau CB, Mihasan M, Ganas P, Igloi GL, Artenie V, Brandsch R (2006) Final steps in the catabolism of nicotine. FEBS J 273:1528–1536
Civilini M, Domenis C, Sebastianutto N, de Bertoldi M (1997) Nicotine decontamination of tobacco agro-industrial waste and its degradation by micro-organisms. Waste Manag Res 15:349–358
dePalmenaer D, Siguier P, Mahillon J (2008) IS4 family goes genomic. BMC Evol Biol 8:18
Ganas P, Sachelaru P, Mihasan M, Igloi GL, Brandsch R (2008) Two closely related pathways of nicotine catabolism in Arthrobacter nicotinovorans and Nocardioides sp. strain JS614. Arch Microbiol 189:511–517
Ganas P, Igloi GL, Brandsch R (2009) The megaplasmid pAO1 of Arthrobacter nicotinovorans and nicotine catabolism. In: Steinbüchel A, Schwartz E (eds) Microbial megaplasmids. Springer Berlin Heidelberg, Germany, pp 271–282
Gherna RL, Richardson SH, Rittenberg SC (1965) The bacterial oxidation of nicotine. VI. The metabolism of 2,6-dihydroxypseudooxynicotine. J Biol Chem 240:3669–3674
Gladyshev VN, Khangulov SV, Stadtman TC (1994) Nicotinic acid hydroxylase from Clostridium barkeri: electron paramagnetic resonance studies show that selenium is coordinated with molybdenum in the catalytically active selenium-dependent enzyme. Proc Natl Acad Sci U S A 91:232–236
Heeb S, Blumer C, Haas D (2002) Regulatory RNA as mediator in GacA/RsmA-dependent global control of exoproduct formation in Pseudomonas fluorescens CHA0. J Bacteriol 184:1046–1056
Herrero M, de Lorenzo V, Timmis KN (1990) Transposon vectors containing non-antibiotic resistance selection markers for cloning and stable chromosomal insertion of foreign genes in gram-negative bacteria. J Bacteriol 172:6557–6567
Hurh B, Yamane T, Nagasawa T (1994) Purification and characterization of nicotinic acid dehydrogenase from Pseudomonas fluorescens. J Ferment Bioeng 78:19–26
Igloi GL, Brandsch R (2003) Sequence of the 165-kilobase catabolic plasmid pAO1 from Arthrobacter nicotinovorans and identification of a pAO1-dependent nicotine uptake system. J Bacteriol 185:1976–1989
Kachalova GS, Bourenkov GP, Mengesdorf T, Schenk S, Maun HR, Burghammer M, Riekel C, Decker K, Bartunik HD (2010) Crystal structure analysis of free and substrate-bound 6-hydroxy-l-nicotine oxidase from Arthrobacter nicotinovorans. J Mol Biol 396:785–799
Keen NT, Tamaki S, Kobayashi D, Trollinger D (1988) Improved broad-host-range plasmids for DNA cloning in gram-negative bacteria. Gene 70:191–197
Laia ML, Moreira LM, Dezajacomo J, Briati JB, Ferreria CB, Ferro M, Silva A, Ferro J, Oliveira J (2009) New genes of Xanthomonas citri subsp. citri involved in pathogenesis and adaption revealed by a transposon-based mutant library. BMC Microbiol 9:9–12
Li H, Xie K, Huang H, Wang S (2014) 6-Hydroxy-3-succinoylpyridine hydroxylase catalyzes a central step of nicotine degradation in Agrobacterium tumefaciens S33. PLoS ONE 9(7):e103324
Liu Y, Wang L, Huang K, Wang W, Nie X, Jiang Y, Li P, Liu S, Xu P, Tang H (2014) Physiological and biochemical characterization of a novel nicotine-degrading bacterium Pseudomonas geniculata N1. PLoS ONE 9:e84399
Mihasan M, Chiribau CB, Friedrich T, Artenie V, Brandsch R (2007) An NAD (P) H-nicotine blue oxidoreductase is part of the nicotine regulon and may protect Arthrobacter nicotinovorans from oxidative stress during nicotine catabolism. Appl Environ Microbiol 73(8):2479–2485
Nagel M, Andreesen JR (1990) Purification and characterization of the molybdoenzymes nicotinate dehydrogenase and 6-hydroxynicotinate dehydrogenase from Bacillus niacini. Arch Microbiol 154:605–613
Novotny TE, Zhao F (1999) Consumption and production waste: another externality of tobacco use. Tob Control 8:75–80
Qiu J, Ma Y, Wen Y, Chen L, Wu L, Liu W (2010) Functional identification of two novel genes from Pseudomonas sp. strain HZN6 involved in the catabolism of nicotine. Appl Environ Microbiol 78:2154–2160
Qiu J, Ma Y, Zhang J, Wen Y, Liu W (2013) Cloning of a novel nicotine oxidase gene from Pseudomonas sp. strain HZN6 whose product nonenantioselectively degrades nicotine to pseudooxynicotine. Appl Environ Microbiol 79:2164–2171
Qiu J, Wei Y, Ma Y, Wen R, Wen Y, Liu W (2014) A novel (S)-6-hydroxynicotine oxidase gene from Shinella sp. strain HZN7. Appl Environ Microbiol 80(18):5552–5560
Reznikoff WS, Winterberg KM (2008) Transposon-based strategies for the identification of essential bacterial genes. Methods Mol Biol 416:13–26
Sachelaru P, Schiltz E, Igloi GL, Brandsch R (2005) An α/β-fold C-C bond hydrolase is involved in a central step of nicotine catabolism by Arthrobacter nicotinovorans. J Bacteriol 187:8516–8519
Sachelaru P, Schiltz E, Brandsch R (2006) A functional mobA gene for molybdopterin cytosine dinucleotide cofactor biosynthesis is required for activity and holoenzyme assembly of the heterotrimeric nicotine dehydrogenases of Arthrobacter nicotinovorans. Appl Environ Microbiol 72:5126–5131
Sambrook J, Fritsch EF, Maniatis T (1989) Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory, Cold Spring Harbor
Schäfer A, Tauch A, Jäger W, Kalinowski J, Thierbach G, Pühler A (1994) Small mobilizable multi-purpose cloning vectors derived from the Escherichia coli plasmids pK18 and pK19: selection of defined deletions in the chromosome of Corynebacterium glutamicum. Gene 145:69–73
Schenk S, Hoelz A, Krauss B, Decker K (1998) Gene structures and properties of enzymes of the plasmid-encoded nicotine catabolism of Arthrobacter nicotinovorans. J Mol Biol 284:1323–1339
Sloan FA, Gelband H (2007) Cancer control opportunities in low- and middle-income countries. Institute of Medicine (U.S.), Committee on Cancer Control in Low- and Middle-Income Countries, National Academies Press, Washington DC
Tamura K, Dudley J, Nei M, Kumar S (2007) MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol Biol Evol 24:1596–1599
Tang H, Wang L, Meng X, Ma L, Wang S, He X, Wu G, Xu P (2009) Novel nicotine oxidoreductase-encoding gene involved in nicotine degradation by Pseudomonas putida strain S16. Appl Environ Microbiol 75:772–778
Tang H, Yao Y, Wang L, Yu H, Ren Y, Wu G, Xu P (2012) Genomic analysis of Pseudomonas putida: genes in a genome island are crucial for nicotine degradation. Sci Rep 2:377
Tang H, Wang L, Wang W, Yu H, Zhang K, Yao Y, Xu P (2013) Systematic unraveling of the unsolved pathway of nicotine degradation in Pseudomonas. PLoS Genet 9:e1003923
Thomas WJ, Thireault CA, Kimbrel JA, Chang JH (2009) Recombineering and stable integration of the Pseudomonas syringae pv. syringae 61 hrp/hrc cluster into the genome of the soil bacterium Pseudomonas fluorescens Pf0-1. Plant J 60:919–928
Wada E, Yamasaki K (1953) Mechanism of microbial degradation of nicotine. Science 117:152–153
Wang S, Huang H, Xie K, Xu P (2012) Identification of nicotine biotransformation intermediates by Agrobacterium tumefaciens strain S33 suggests a novel nicotine degradation pathway. Appl Microbiol Biotechnol 95(6):1567–1578
Wei HL, Zhang LQ (2006) Quorum-sensing system influences root colonization and biological control ability in Pseudomonas fluorescens 2P24. Anton Leeuw Int J G 89:267–280
Wei HL, Lei LP, Xia ZY, Liu XZ (2008) Characterization of a novel aerobic nicotine-biodegrading strain of Pseudomonas putida. Ann Microbiol 58:41–45
Wei HL, Lei L, Liu S, Xia Z, Liu X, Liu P (2009) PanB is involved in nicotine metabolism in Pseudomonas putida. Int Biodeterior Biodegrad 63:988–992
Yao Y, Tang H, Ren H, Yu H, Wang L, Zhang W, Behrman EJ, Xu P (2013) Iron (II)-dependent dioxygenase and N-formylamide deformylase catalyze the reactions from 5-hydroxy-2-pyridone to maleamate. Sci Rep 3:3235
Yu H, Tang H, Wang L, Yao Y, Wu G, Xu P (2011) Complete genome sequence of the nicotine-degrading Pseudomonas putida strain S16. J Bacteriol 193:5541–5542
Yu H, Li Y, Tang H, Xu P (2014) Genome sequence of a newly isolated nicotine-degrading bacterium, Ochrobactrum sp. SJY1. Genome Announc 2(4):e00720-14
Acknowledgments
This work was supported by a grant from the International Foundation for Science (F/4583-1) to Hai-Lei Wei and a grant from the National Natural Science Foundation of China (30760011) to Liping Lei.
Conflict of interest
The authors have no conflict of interest to declare.
Author information
Authors and Affiliations
Corresponding author
Additional information
Zhenyuan Xia and Wei Zhang contributed equally to this work.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Table S1
(PDF 38 kb)
Rights and permissions
About this article
Cite this article
Xia, Z., Zhang, W., Lei, L. et al. Genome-wide investigation of the genes involved in nicotine metabolism in Pseudomonas putida J5 by Tn5 transposon mutagenesis. Appl Microbiol Biotechnol 99, 6503–6514 (2015). https://doi.org/10.1007/s00253-015-6529-x
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-015-6529-x