Abstract
Almost half the human genome consists of mobile DNA elements, and their analysis is a vital part of understanding the human genome as a whole. Many of these elements are ancient and have persisted in the genome for tens or hundreds of millions of years, providing a window into the evolution of modern mammals. The Golem family have been used as model transposons to highlight computational analyses which can be used to investigate these elements, particularly the use of molecular dating with large transposon families. Whole-genome searches found Golem sequences in 20 mammalian species. Golem A and B subsequences were only found in primates and squirrel. Interestingly, the full-length Golem, found as a few copies in many mammalian genomes, was found abundantly in horse. A phylogenetic profile suggested that Golem originated after the eutherian–metatherian divergence and that the A and B subfamilies originated at a much later date. Molecular dating based on sequence diversity suggests an early age, of 175 Mya, for the origin of the family and that the A and B lineages originated much earlier than expected from their current taxonomic distribution and have subsequently been lost in some lineages. Using publically available data, it is possible to investigate the evolutionary history of transposon families. Determining in which organisms a transposon can be found is often used to date the origin and expansion of the families. However, in this analysis, molecular dating, commonly used for determining the age of gene sequences, has been used, reducing the likelihood of errors from deleted lineages.
Similar content being viewed by others
Background
Mobile DNA sequences form a major component of the mammalian genome, and comprise, for example, 44% of the human genome (Mills et al. 2007). A family of transposable elements is most typically regarded as a parasitic entity, which is capable, through its over-replication relative to host DNA, of increasing its numbers, even if this increase can bring a reduction in fitness to the hosts. In this process, the family itself may evolve into autonomous and non-autonomous elements, the latter being capable of transposition only in cells with active autonomous elements of the same family, which are capable of supplying the trans-acting components of transposition. In such a situation the non-autonomous elements can be seen as analogous to hyper-parasites.
The genomes of higher organisms differ greatly in the rate at which non-functional DNA sequences are removed. In Drosophila, it appears that almost all the non-coding DNAs are subject to purifying selection, and any non-coding sequences that lack function tend to be eliminated (with a half-life of perhaps 14 million years) (Halligan and Keightley 2006; De Proce et al. 2009; Petrov and Hartl 1998). However, in other groups, such as the mammals and the flowering plants, functionless DNA sequences appear to be able to persist, with the consequence that the relics of mobile sequence families active tens of millions of years ago can still be identified and studied (Xie et al. 2006).
What kind of selection will act on transposable element sequences? If mobile DNAs are purely parasitic sequences which produce no benefits for their hosts, then, at any given chromosomal location, inactivating mutations will be neutral with respect to natural selection. Indeed, such inactivating mutations could be weakly advantageous as they will reduce the rate at which a mobile DNA sequence produces potentially harmful daughter elements. The consequence of this is that we would expect the copies of a mobile sequence family, located at an individual chromosomal position, to evolve at a rate equal to (or perhaps very slightly greater than) the mutation rate. This would be likely to result in any element, located in its current chromosomal position for millions of years, losing any cis-acting sequences required for its transposability.
We can think of these processes as the life cycle of a mammalian transposable element family in which, initially and probably as a result of a horizontal transfer (Pace et al. 2008), the element appears and proliferates in the genome. It creates hundreds or thousands of daughter elements, many of which become inactive as a result of mutations. A small subset of active elements remains, continuing to transpose and, as a result, continue to be subject to purifying selection. Over time, for element families that are capable of mutating to diversify into autonomous and non-autonomous forms, the spread of the non-autonomous forms may, under some restricted circumstances, drive the autonomous forms to extinction (Brookfield 1991, 1996). Eventually, both the autonomous and non-autonomous forms may all become inactive as, in a given lineage, the trans-acting functions of autonomous elements become subject to inactivating mutations. In mammalian genomes, which do not clear away their functionless DNAs, the remains of this process are still visible to genomic archaeologists.
In the study of this process of spread and inactivation of a family, we are helped by the fact that the process lasts for many millions of years, during which time the lineage can split into what are today very diverse descendant lineages. This gives us more power to reconstruct ancestral events through the pooling of information from multiple extant lineages, but also allows us to see whether the process of proliferation, evolution and inactivation differed between the various lineages into which the initially invaded genome subsequently diversified.
The proliferation of available genome sequences for mammalian organisms allows the collation and analysis of large transposon families using bioinformatics techniques. Traditionally, these families have been dated by phylogenetic analysis, either through similarity to other transposons or by an analysis of which organisms the elements can be found in. We suggest the use of molecular dating using the Markov Chain Monte Carlo methods BEAST (Drummond and Rambaut 2007) and PhyloBayes 3 (Lartillot et al. 2009) and show that this method can help to decide between possible hypotheses suggested by the phylogenetic analysis.
Here, we investigate the active phase of the Class II element Golem (Smit and Riggs 1996; Paulis et al. 2004), and its non-autonomous relations (Golem A and Golem B), in mammalian genomes. Golem was chosen as a model transposable element, in which to test these techniques, primarily because it appeared to be a typical class II mammalian transposable element with a large number of sequences still identifiable in the genomes of modern mammals. A large number of similar elements exist, such as the Tigger elements and many other mariner-like elements, it is expected that techniques which work well for the Golem sequences could be used to investigate these similar transposons. Further to these reasons, Golem was of interest to us due to its two deletion products allowing an investigation into when these shorter elements first appeared and how widespread they are across mammalian species. While many class II transposons have a related deletion product the appearance of two, both of which appear to have maintained an ability to transpose, despite the loss of the internal ORF sequence, is relatively unusual.
Golems A and B (Fig. 1) appear to have been the result of large deletions in the original sequence which were then propagated as new members of the Golem family. These subsequences do not contain ORFs which would allow the autonomous transposition of the sequences; however, their propagation throughout the genomes implies that the transposition is occurring, most likely through another agent such as the original Golem sequence. We see the ways in which it has spread and been inactivated, and the ways in which this latter process has differed between different host species.
Results and Discussion
The Overall Structure of the Family
The Golem family of transposon sequences could only be identified in a subset of mammalian genomes. Blast searches resulted in Golem matches in genomes from 20 organisms; however, Golem A- and Golem B-specific sequences were only found in primates and to a very small extent in rodentia genomes (Fig. 2). The occurrence of Golem sequences in some organisms, but not in other phylogenetically related species, implies that Golem may have been deleted in certain organisms while retained in others. The existence of multiple copies of Golem in the horse (Equus caballus) genome, where none exist in the cow (Bos taurus) or pig (Sus scrofa) genomes, is an example of this retention. An alternative hypothesis is that, perhaps, only one or a few copies existed in the horse ancestor, and the existence of the numerous sequences in the horse genome is due to a later resurgence in this species. To confirm that this occurrence of Golem in the horse genome, but not in cow or pig, was not a symptom of a wider issue, a brief examination of the pre-masked genomes in repeat masker (http://www.repeatmasker.org) was undertaken. No significant differences can be seen in the percentage of the genomes that consists of transposons or that consist of DNA transposons.
Golem family frequencies in mammals. Barcharts showing the number of sequences matching each of the Golem-, Golem A- and Golem B-specific regions in all organisms which returned any matches to the Golem query sequences. Organisms queried against the NCBI genome database can be identified by (G). Golem-, Golem A- or Golem B-specific values show the number of sequences with matches to the diagnostic sequences for these transposon subfamilies. The 75% Golem, Golem A or Golem B values show the number of sequences which showed similarity to ≥75% of the Golem, Golem A or Golem B consensus sequence
These Golem sequences found in the horse genome thus exhibit an unusual distribution. A large number of Golem sequences spanning the entire length of the consensus, including the internal ORF, can still be found in the genome (Fig. 1). All examples of the ORF have been found to contain either stop codons or frameshift mutations, implying that no actively transposing Golem sequences are still present. Examples of similar full-length sequences can be found in primates; however, these are found in much smaller numbers. Of these primate full-length Golem sequences, none were found with both homology and synteny to the full-length sequences found in the horse genome. This observation may lend weight to the theory that a resurgence in Golem activity occurred in the horse genome. However, it may simply be that Golem was still actively transposing after the horse–human divergence and that sequences inserted into the genome before this time point have now diverged to such an extent to be no longer recognisable as homologues.
Where synteny can be reliably established, in primates, the elements that are most similar in sequence are also shown to be orthologous from their genomic locations (Fig. 3). The implication is that gene conversion has played a comparatively small role in the recent evolution of the sequences. It can also be seen from the non-human primate sequences, orthologous to human Golem transposon copies, that Golem had ceased to be active prior to the human–chimpanzee split and the inactivation was probably early in the primate lineage.
The Family Shows Only Inconsistent Evidence of Having Acquired a Function at the Level of the Host
Orthologous copies of a mobile DNA sequence are expected to diverge at the basal mutation rate as mutations in such sequences, including inactivating mutations, will not lower host fitness. Thus, if individual copies show sequence conservation relative to the neutral evolutionary rate, this can be taken as evidence for “domestication”, in which that individual element copy is involved in a sequence-dependent function that is adaptive at the level of the host.
We examined the distribution of divergences in human–chimpanzee pairs. Of 3,163 human–chimpanzee orthologous pairs identified, 254 were identical in sequence. However, it cannot be concluded that this identity implies conservation, since the evolutionary divergence of the two species is so small that short, neutrally evolving, DNA sequences will often be identical. If “domesticated” sequences exist among those showing low divergence, one would expect a divergence in evolutionary rate between copies.
We can predict an expected distribution of sequence variation between human–chimpanzee orthologues, assuming that the number of substitutions between the sequences is Poisson distributed with a mean equal to the overall sequence divergence, multiplied by the length of that element. The observed variance (0.875646) in percentage sequence divergence is significantly greater than that expected (0.444289) from the assumption that all copies have the same evolutionary rate. This is consistent with some copies being subject to purifying selection. However, we noted that some of the pairs showed very high divergences in sequence (>5%) which argues that they may not be true orthologues, but rather have been affected by gene conversions, for example. Also, it has been noted (Patterson et al. 2006) that there is heterogeneity in human–chimpanzee sequence divergence between different genomic regions, which may reflect divergence in times to common ancestry. This would be expected to inflate the observed variance.
In order to avoid the effects of highly diverged sequences that may not be true orthologues, we considered only the 254 identical sequences and calculated the number expected to be identical based on the genome-wide nucleotide divergence of 1.23% (Mikkelsen and The chimpanzee Sequencing and Analysis Consortium 2005). For each of the sequences, we calculated the Poisson probability that it would be identical in the two species as exp(−0.0123 × length), where “length” is the length of that particular sequence. This analysis predicts only 151 identical sequence pairs. Simulations revealed a probability <0.00001 of seeing as many as 254 identical pairs. The observation of 254 provides significant evidence (P = 0.05) against a rate over 1.01% and against a rate under 0.92%.
However, there is variation in human–chimpanzee divergence between genomic regions (Patterson et al. 2006), and the more variation there is, the higher is the expected number of sequences with zero divergence. Patterson et al. described a range of times to human–chimpanzee common ancestry for different parts of the genome which ranged from 84 to 147% of the overall average. Thus, we carried out simulations in which, on average, 25.4% of the sequences had times to common ancestry sampled from a flat distribution in the range from 100 to 147% of the mean of 1.23% divergence, and 74.6% of the sequences sampled from a flat distribution between 84 and 100% of the mean. The 25.4:74.6 ratio was chosen since this predicted a divergence of 100% of the observed mean overall. This incorporation of between-element divergence time only had the effect of raising the expected number of identical sequence pairs to 160, and seeing 254 again had a probability below 0.00001.
We have identified 1,000 nucleotides upstream and downstream of all elements in the human Golem dataset. Comparing these with chimpanzee orthologues reveals a mean divergence of 0.0157. Simulations using this divergence again yield a probability of 254 or more sequences identical to be below 0.00001.
This observation of an unexpectedly high number of identical sequence pairs is consistent with purifying selection, and thus a functional constraint, on some copies of Golem. If some copies are subject to purifying selection and if the selection is consistent across the primates, we would expect there to be a strong correlation between the divergence of a given orthologous element in a human–chimpanzee comparison and the divergence of that same element in, for example, a human–orangutan comparison. We thus focussed on 154 elements which have orthologues in orangutan, macaque and marmoset and observed the correlation between the human–chimpanzee divergence and the divergence between human and the other primate genomes examined. The observed Pearson correlations with the human–chimpanzee divergence, 0.1556 for human–orangutan, 0.1796 for human–macaque and 0.094 for human–marmoset, were compared with the expected values (resulting from the sharing of the branch between human and human–chimpanzee ancestor) in simulations based on the variance in the sequence length and incorporating the variances in divergence across sequences for all species pairs. In no case was the observed correlation higher than the expectation, arguing against a purifying selection that is conserved across the primates, although not against a purifying selection in humans and chimpanzees that differs from that in other groups.
The Golem sequences were analysed for composition and positional bias. No difference in nucleotide composition or CpG depletion could be detected between the Golem sequences and their flanking regions, and there was no positional bias detected with most sequences falling in non-coding DNA which did not appear to be part of promoter regions. We are also confident that this is not an artefact cause by selection bias from the BLAT search methods as altering the sensitivity of the search parameters did not alter the increase in the number of Golem sequences found. However, we would like to note that there may still be further biases, causing this effect, that we have not yet discovered.
Purifying Selection on the Transposable Element’s Predicted Amino Acid Sequence can be Detected at Some, but not all, Phases of Its Evolutionary Existence
Golem was predicted to contain an ORF from positions 463–2,307 of the consensus sequence. The conserved domains searched showed similarity to CENP-B_N (E-value: 9.99e−4) and CENPB (E-value: 4.27e−8), Transposase_Tc5 (E-value: 1.21e−9) and DDE (E-value: 1.4e−48). This pattern of conserved domains can be found in other members of the pogo superfamily, such as the tigger transposon (Kipling and Warbuton 1997). A blast search showed a match to the human tigger transposable element-derived protein 1 (NCBI: NP_663748.1) with an E-value of 1e−133.
Through the use of the full-length Golem sequences, it is possible to use evidence of evolutionary constraint on coding sequences to identify when, in the life of the Golem sequence, purifying selection was or was not operating. Figure 4 shows part of the phylogeny of Golem sequences from the horse genome, where lines in black, showing the time from the most recent known convergences to the sequence tips, are expected to show less constraint in the sequence. Many of these lineages will only include elements that are transpositionally inactive, but our incomplete sampling of the results of transpositions during this time (due to subsequent element losses from the genome) will mean that some of these branches will also represent elements transpositionally active for the early part of the branch. Those lines in grey, connecting internal nodes to the most recent common ancestor, represent the family during its proliferation phase when selection on the open reading frame of the transposase protein would be expected.
Horse Golem phylogeny. A section of the maximum likelihood phylogenetic tree showing the evolutionary history of Golem sequences in the horse genome. Only sequences with >75% coverage of the consensus sequence were included. Only sites with 90% coverage were included in the analysis. Bootstrap values were calculated using 200 replications
Figure 5 shows the ratio of non-synonymous changes to synonymous changes. It can be seen that selective constraint was working to a greater extent in earlier sequences than in was in the later, largely inactivated, sequences. T-tests show a significant difference in d N/d S ratio between the early branches (root to convergence sequence) and the tip to convergence (P < 0.005).
Predicting the Age of the Golem Family
The occurrence of Golem sequences across species and orders was analysed to determine likely points of origin for each of the Golem family members. The full Golem sequence, being found in most placental mammalian species, but not in marsupial species, can be dated to the divergence point between Eutheria and Metatheria. The Arnason estimates of molecular divergences place this time point between 120 and 140 Mya (Arnason et al. 2008). Other studies, however, have dated this as a much earlier event at approximately 180 Mya (Kumar and Hedges 1998; Kumar and Subramanian 2002; Woodburne et al. 2003). The Golem A and B sequences, only found in primates and to a small extent in rodentia, are assumed to have originated at a much later time point approximately 70–100 Mya according to the estimates in the literature (Arnason et al. 2008; Douzery et al. 2003).
BEAST and PhyloBayes 3 are Markov Chain Monte Carlo methods of molecular dating, designed to be used on allelic variation. Although the primate Golem variations are not allelic, we hypothesise that these programs can be used to determine an early date at which the transposon was active. In this example, the bifurcations in the tree near the tip correspond to the splits between primate sequences known to be orthologues and these can be constrained to the date of known speciation events. However, the early branches correspond to bifurcations in element lineages at different genomic sites as the element proliferated through the ancestral genome and which can be seen as a population of elements. By using known primate speciation times to date the bifurcations near the tip, molecular dating techniques can be used to date the early branches and the age of the MRCA of the Golem sequences, a time point assumed to be similar to the origin of the transposon. Although, ideally, this would be the date of the very first active transposon, it cannot be guaranteed that the descendents visible in modern genomes are not all products of a later active transposon.
The MCMC analyses of the Golem sequences in primates, shown in Fig. 6, indicated an age of between 140 and 185 Myr ago, with an overall mean of 151 Myr ago, for the time to common ancestry of sequence copies, which, assuming an invasion, would follow soon after the origin of the family. Both the BEAST analysis, with strict or uncorrelated molecular clocks, and the PhyloBayes 3 analysis approximately agreed on the predicted dates, although the strict clock gave smaller errors and a much higher ESS rate. The human–chimp and human–orangutan speciation events also gave consistent predictions with no consistent difference between results obtained using the human–orangutan date rather than the human–chimp date. A mean mutation rate of 9.5e−4 per base per million years was calculated using the relaxed clock and a slightly lower rate of 9.1e−4 using the strict clock. The difference in the rates was not found to be significant. The horse population of Golem sequences was compared with the primates by setting the mutation rates of both populations as 1 in a BEAST analysis. No evidence was found that the horse population was less diverse, and therefore younger, than the primate populations. This suggests that the resurgence hypothesis is not correct and that loss of Golem in other species is more likely.
MCMC molecular dating of Golem transposons. Bars represent the results of each Beast or PhyloBayes 3 analysis from the lower 95% bound to the upper 95% bound, and circular points represent the mean predicted age of most recent common ancestor. Analysis labelling: C chimpanzee–human time points (mean of 6 Myr ago, standard deviation of 0.5), O chimpanzee–orangutan time points (mean of 13 million years ago, standard deviation of 1), 1 BEAST analysis with uncorrelated log-normal clock, 2 BEAST analysis with strict clock, 3 PhyloBayes analysis with strict clock
Molecular dating of the Golem B sequences places the origin of these sequences at approximately the same time as the Golem sequences. Analyses using the two clocks both give a predicted age of 152 Mya; however, as with the Golem sequences, the error obtained using the strict clock is much lower than that using the relaxed clock. In contrast to these results, Golem A is dated as a much younger member of the Golem family with a mean age of 110 Myr. Both Golem A and Golem B sequences gave a MRCA date much earlier than would be expected from the species in which the sequences are found. This may indicate that the sequences were lost from some lineages while kept in others. Mutation rates for these sequences were consistent with those of Golem (Golem A: 1.9e−3, Golem B: 9.5e−4).
It was hypothesised that the autonomous Golem sequences may be mutating at a slower rate than the non-autonomous sequences due to selective constraints; however, no significant difference can be seen between the mutation rates of the subfamilies (t-test; P > 0.05) and the mutation rates were found to be approximately similar to those that would be expected for non-coding DNA (Kumar and Subramanian 2002).
Conclusions
The Golem family appears to have become active approximately 140–185 Myr ago. From the lack of Golem sequences found in marsupial genomes (Monodelphis domestica, Macropus eugenii), it is reasonable to assume that the origin of Golem occurred soon after the metatherian–eutherian divergence. Although this is early for the metatherian–eutherian divergence by some estimates (Arnason et al. 2008; Bininda-Emonds et al. 2008), other analyses have dated the divergence to within a reasonable margin of error from this date (Kumar and Hedges 1998; Kumar and Subramanian 2002; Woodburne et al. 2003).
Although the Golem A and Golem B lineages only appear in primate and rodentia species, the molecular dating places their origin dates much earlier than would be assumed from this pattern of occurrence. The dates would, by most estimates, imply that the origin of Golem B occurred at a time of common ancestry for most eutherian organisms. The predicted origin date of Golem A, while later than Golem B, would still suggest that it was found in the common ancestor of many more species than it is found in today. A possible explanation for the discrepancy between predicted origin dates and the lack of Golem A and Golem B in most mammalian orders is that the sequences existed in small numbers until just before the primate divergence. Following the divergence of these species, the sequences increased rapidly in the primate common ancestor genome; however, they were lost from other organisms. A further possibility is that the mutation rates of the transposon sequences have slowed as they have become inactivated, possibly due to a high error rate in the transposition process, and this would increase the age of insertion as the constrained dates were assigned to inactivated transposons.
While the inactivation date of Golem has not been completely defined, it is clear from the primate phylogenies that most of the human Golem sequences have synteny and homology to sequences in the other primate species, both Old and New World. Therefore, it is reasonable to assume that most, if not all, Golem sequences were inactive by the time of the old world–new world primate divergence. A small number of Golem sequences may, however, have still been active at this time. Novel sequences can be found in marmosets, and for many human sequences, no homologues can be found. Alternatively, this may be due to deletion rather than the formation of new transposon sequences.
The horse phylogeny shows some lineages of Golem continuing to replicate until quite recently. The lack of a conserved ORF implies that the sequences are now inactive; however, the inactivation date may be much later in this species than in primates. If genomes phylogenetically related to the horse were to become available, a comparison of orthologous Golem sequences would enable an inactivation date to be suggested.
The molecular analysis of transposon sequences can give another avenue into the exploration of the timescale and evolution of the genomes of both human and other organisms. Through the use of the Golem transposons, and its deletion products, as a model system we have tested the migration of several population genetics tools into this field. We have found significant differences between the dates suggested through phylogenetic analysis and molecular dating and would suggest that our method may be compensating for the deletion of transposon sequences early in a species lineage, an event which seriously compromises the dating of transposon sequences through phylogenetic comparison. However, the analysis relies heavily on the existence of genomes with high-level coverage. While genes may be studied using only 2× coverage, the analysis of non-coding DNA, and the discoveries available through such, is much better served using genomes with high-level coverage.
Methods
Identification of Golem Sequences in Mammalian Species
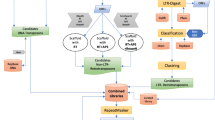
Consensus sequences were retrieved from Repbase Update (Jurka et al. 2005) for Golem, Golem A and Golem B. The NCBI Genomes database (Sayers et al. 2010) was queried with the consensus sequences using Megablast (Zhang et al. 2000). Where more than one genome assembly was available, only sequences from the primary reference assembly were included. Further mammalian genomes, with lower coverage or less complete annotation, were queried using the Ensembl BLAT search (Kent 2002).
Sequences matching the internal sequence of Golem, which are deleted in both Golem A and B, were classified as Golem sequences. Those containing the specific deletions shown in Golem A or B were categorised into these subfamilies and partial sequences which matched regions shared by more than one type of Golem sequence were classified as unknown.
Comparing Golem Sequences Within Species
Creating multiple sequence alignments (MSA) of the sequences using a traditional MSA alignment algorithm, such as ClustalW (Thompson et al. 1994), was problematic due to the number, degeneracy and varying lengths of the sequences. Instead, an assembly algorithm was implemented, using default settings, through the Genious software package (http://www.geneious.com). The Repbase consensus sequences were used as references. Motifs retrieved using the Golem, Golem A and Golem B reference sequences were assembled separately.
Sequences which appeared to be more closely related than expected were investigated, particularly pairs of sequences located on the same chromosome. Sequences 5000 bp up- and downstream from the Golem motif were extracted and locally aligned. The similarity between the Golem motifs, the left flanks and the right flanks at each of the locations were calculated. The opposite flanking sequences (e.g. Sequence 1 right flank vs. Sequence 2 left flank) were compared as a negative control. Golem motifs with at least one of the flanking regions showing significantly greater similarity than seen in the controls were assumed to have occurred as part of a larger duplication, rather than as a transposition event, and one copy was removed from the data set.
Mega 4 (Tamura et al. 2007) was used to create individual phylogenies for the Golem, Golem A and Golem B sequences. Phylogenies were created using neighbour joining with pairwise deletions after removing partial sequences where ≥25% of the sequence was lost.
Comparing Golem Sequences Between Species
Each human Golem sequence was used as the query for a UCSC BLAT search against the Golem sequences from other primate species (Pan troglodytes, Pongo pygmaeus, Macaca mulatta and Callithrix jacchus). The highest scoring match was assumed to be a homologue. Synteny between the homologues was checked though Ensembl, although data were not available for all Golem sequences. The sequences were used to create primate phylogenies for Golem, Golem A and Golem B.
Analysis of Functional Relevance of the Golem Motif
The percentage similarity between human Golem, Golem A and Golem B sequences and their homologous primate sequences was calculated using UCSC BLAT for each of 3,163 human Golem sequences where the orthologous sequences could be established. Variance in similarity expected for a Poisson distribution was calculated and compared to the observed variance. Simulations were carried out using bespoke programs written in C++.
Analysis of Selection on Full-Length Golem Sequences
A NCBI ORF finder (http://www.ncbi.nlm.nih.gov/projects/gorf/) search was conducted to determine the likelihood and position of an internal ORF in the Golem sequence. An NCBI conserved domain search was carried out on the predicted ORF to determine whether the features required of a class II transposase were present. A comparison of the rates of synonymous and non-synonymous mutations (Kimura 1977; Yang and Bielawski 2000) in this region was carried out to determine whether the ORF had been under purifying selection. Frame shifting mutations were removed from the ORF sequences of the full-length Golem alignments. The consensus sequence between each pair of Golem sequences was calculated (which we refer to as “convergence sequences”), as was the overall consensus sequence, representing the root sequence. Using Mega 4, d N/d S was calculated between each Golem sequence and the related convergence sequence, between the Golem sequences and the root sequence, and between the convergence sequences and the root sequence. Student’s t-test was used to determine whether the rates in the three groups were significantly different.
Predicting the Age of the Family
BEAST v1.6.1 (Drummond and Rambaut 2007) and PhyloBayes 3 (Lartillot et al. 2009) were used to date the origin of the Golem family and the appearance of the Golem A and Golem B forms. The analysis was carried out using a MSA consisting of chimpanzee, macaque, orangutan and marmoset sequences orthologous to the 75% coverage Golem, Golem A or Golem B human sequences. Beast analyses were carried out using either a relaxed uncorrelated log-normal molecular clock (Drummond et al. 2006) or a strict clock for each Golem subtype, and PhyloBayes 3 analyses were carried out using a strict clock. The three analyses were carried out to allow for confirmation of the root height and mutation rates and to determine whether a consistent difference could be seen between results obtained from each method. A Yule process tree prior was used in BEAST; however the trees were created in MrBayes (Huelsenbeck and Ronquist 2001; Ronquist and Huelsenbeck 2003) for use with PhyloBayes 3. Each analysis consisted of MCMC runs of 1 Million states, sampled every 1000 states. Initially a 6 Myr time point (normally distributed with a 0.5 standard deviation) for the human–chimpanzee split was used to constrain the analysis. Following this, another analysis was carried out using each of the MCMC methods where the analysis was constrained using an orangutan divergence time point at 13 Myr, with a standard deviation of 1 (Glazko and Nei 2003). Comparisons of the results based on constraining the data at chimpanzee and at orangutan time points will compensate for any effects caused by either a higher than expected level of conservation between the human–chimpanzee or human–orangutan homologues. However, if both sets of homologues are conserved by the same amount, any differences will not be observable in a comparison of the mutation rates or root heights.
To predict the root age of the Golem sequences found in the horse genome, an alternative method was used as no homologous sequences to the horse Golem sequences could be identified. The full-length Golem populations found in horse, human, chimpanzee and orangutan were analysed separately using BEAST. No time constraints were used; instead, the mutation rate was set to 1. The root height was, therefore, measured in mutations/site. The standardisation of the mutation rate allows for a comparison of the predicted root heights between the populations. The method assumes that there is no significant inter-species difference in mutation rates of non-coding DNA.
References
Arnason U, Adegoke JA, Gullberg A, Harley EH, Janke A, Kullberg M (2008) Mitogenomic relationships of placental mammals and molecular estimates of their divergences. Gene 421:37–51
Bininda-Emonds O, Cardillo M, Jones KE, MacPhee RD, Beck RM, Grenyer R, Price SA, Vos RA, Gittleman JL, Purvis A (2008) The delayed rise of present-day mammals. Nature 446:507–512
Brookfield JFY (1991) Models of repression of transposition in P-M hybrid dysgenesis by P-cytotype and by zygotically-encoded repressor proteins. Genetics 128:471–486
Brookfield JFY (1996) Models of the spread of non-autonomous selfish transposable elements when transposition and fitness are coupled. Genet Res 67:199–209
De Proce SM, Halligan DL, Keightley PD, Charlesworth B (2009) Patterns of DNA sequence divergence between Drosophila miranda and D. pseudoobscura. J Mol Evol 69:601–611
Douzery EJ, Delsuc F, Stanhope MJ, Huchon D (2003) Local molecular clocks in three nuclear genes: divergence times for rodents and other mammals and incompatibility among fossil calibrations. J Mol Evol 57(Suppl 1):201–213
Drummond AJ, Rambaut A (2007) BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol Biol 7:214
Drummond AJ, Ho SYW, Phillips MJ, Rambaut A (2006) Relaxed phylogenetics and dating with confidence. PLoS Biol 8:114
Glazko GV, Nei M (2003) Estimation of divergence times for major lineages of Primate species. Mol Biol Evol 20:424–434
Halligan DL, Keightley PD (2006) Ubiquitous selective constraints in the Drosophila genome revealed by a genome-wide interspecies comparison. Genome Res 16:875–884
Huelsenbeck JP, Ronquist F (2001) MRBAYES: Bayesian inference of phylogeny. Bioinformatics 17:754–755
Jurka J, Kapitonov VV, Pavlicek A, Klonowski P, Kohany O, Walichiewicz J (2005) Repbase update, a database of eukaryotic repetitive elements. Cytogenet Genome Res 110:462–467
Kent WJ (2002) BLAT—the BLAST-like alignment tool. Genome Res 12:656–664
Kimura M (1977) Preponderance of synonymous changes as evidence for the neutral theory of molecular evolution. Nature 267:275–276
Kipling D, Warbuton PE (1997) Centromeres, CENP-B and Tigger too. Trends Genet 13(Suppl 4):141–145
Kumar S, Hedges SB (1998) A molecular timescale for vertebrate evolution. Nature 292:917–920
Kumar S, Subramanian S (2002) Mutation rates in mammalian genomes. Proc Natl Acad Sci USA 99:803–808
Lartillot N, Lepage T, Blanquart S (2009) PhyloBayes 3: a Bayesian software package for phylogenetic reconstruction and molecular dating. Bioinformatics 25:2286–2288
Mikkelsen TS, The chimpanzee Sequencing and Analysis Consortium (2005) Initial sequence of the chimpanzee genome and comparison with the human genome. Nature 437:69–87
Mills RE, Bennett EA, Iskow RC, Devine SE (2007) Which transposable elements are active in the human genome? Trends Genet 23:183–191
Pace JK, Gilbert C, Clark MS (2008) Feschotte C: repeated horizontal transfer of a DNA transposon in mammals and other tetrapods. Proc Natl Acad Sci USA 105:17023–17028
Patterson N, Richter DJ, Gnerre S, Lander ES, Reich D (2006) Genetic evidence for complex speciation of humans and chimpanzees. Nature 441:1103–1108
Paulis M, Moralli D, Bensi M, De Carli L, Raimondi E (2004) Isolation from the horse genome of a new DNA transposon belonging to the Tigger family. Mamm Genome 15:399–403
Petrov DA, Hartl DL (1998) High rate of DNA loss in the Drosophila melanogaster and Drosophila virilise species groups. Mol Biol Evol 15:293–302
Ronquist F, Huelsenbeck JP (2003) MRBAYES 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19:1572–1574
Sayers EW, Barrett T, Benson DA, Bolton E, Bryant SH, Canese K, Chetvernin V, Church DM, Dicuccio M, Federhen S, Feolo M, Fingerman IM, Geer LY, Helmberg W, Kapustin Y, Landsman D, Lipman DJ, Lu Z, Madden TL, Madej T, Maglott DR, Marchler-Bauer A, Miller V, Mizrachi I, Ostell J, Panchenko A, Phan L, Pruitt KD, Schuler GD, Sequeira E et al (2010) Database resources of the National Center for Biotechnology Information. Nucleic Acids Res 39:D38–D51
Smit AF, Riggs AD (1996) Tiggers and DNA transposon fossils in the human genome. Proc Natl Acad Sci USA 93(Suppl 4):1443–1448
Tamura K, Dudley J, Nei M, Kumar S (2007) MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol Biol Evol 24:1596–1599
Thompson JD, Higgins DG, Gibson TJ (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res 22:4673–4680
Woodburne MO, Rich TH, Springer MS (2003) The evolution of tribospheny and the antiquity of mammalian clades. Mol Phylogenet Evol 28:360–385
Xie X, Kamal M, Lander ES (2006) A family of conserved noncoding elements derived from an ancient transposable element. Proc Natl Acad Sci USA 103:11659–11664
Yang Z, Bielawski JP (2000) Statistical methods for detecting molecular adaptation. Trends Ecol Evol 15:496–503
Zhang Z, Schwartz S, Wagner L, Miller W (2000) A greedy algorithm for aligning DNA sequences. J Comput Biol 7:203–214
Acknowledgements
We would like to thank the BBSRC (research grant reference: BB/H009884/1) for funding this study.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
Open Access This is an open access article distributed under the terms of the Creative Commons Attribution Noncommercial License (https://creativecommons.org/licenses/by-nc/2.0), which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
About this article
Cite this article
Hellen, E.H.B., Brookfield, J.F.Y. Investigation of the Origin and Spread of a Mammalian Transposable Element Based on Current Sequence Diversity. J Mol Evol 73, 287–296 (2011). https://doi.org/10.1007/s00239-011-9475-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00239-011-9475-y