Abstract
Key message
The rapid accumulation of pre-existing mutations may play major roles in the establishment and shaping of adaptability for local regions in current rice breeding programs.
Abstract
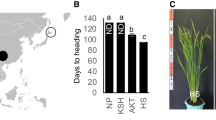
The cultivated rice, Oryza sativa L., which originated from tropical regions, is now grown worldwide due to the concerted efforts of breeding programs. However, the process of establishing local populations and their origins remain unclear. In the present study, we characterized DNA polymorphisms in the rice variety KITAAKE from Hokkaido, one of the northern limits of rice cultivation in the world. Indel polymorphisms were attributed to transposable element-like insertions, tandem duplications, and non-TE deletions as the original mutation events in the NIPPONBARE and KITAAKE genomes. The allele frequencies of the KITAAKE alleles markedly shifted to the current variety types among the local population from Hokkaido in the last two decades. The KITAAKE alleles widely distributed throughout wild rice and cultivated rice over the world. These have accumulated in the local population from Hokkaido via Japanese landraces as the ancestral population of Hokkaido. These results strongly suggested that combinations of pre-existing mutations played a role in the establishment of adaptability. This approach using the re-sequencing of local varieties in unique environmental conditions will be useful as a genetic resource in plant breeding programs in local regions.



Similar content being viewed by others
References
Alonso-Blanco C, Blankestijn-de Vries H, Hanhart CJ, Koornneef M (1999) Natural allelic variation at seed size loci in relation to other life history traits of Arabidopsis thaliana. Proc Natl Acad Sci USA 96:4710–4717
Ammiraju JS, Luo M, Goicoechea JL, Wang W, Kudrna D et al (2006) The Oryza bacterial artificial chromosome library resource: construction and analysis of 12 deep-coverage large-insert BAC libraries that represent the 10 genome types of the genus Oryza. Genome Res 16:140–147
Ammiraju JS, Lu F, Sanyal A, Yu Y, Song X et al (2008) Dynamic evolution of Oryza genomes is revealed by comparative genomic analysis of a genus-wide vertical data set. Plant Cell 20:3191–3209
Arai-Kichise Y, Shiwa Y, Nagasaki H, Ebana K, Yoshikawa H, Yano M, Wakasa K (2011) Discovery of genome-wide DNA polymorphisms in a landrace cultivar of Japonica rice by whole-genome sequencing. Plant Cell Physiol 52:274–282
Arai-Kichise Y, Shiwa Y, Ebana K, Shibata-Hatta M, Yoshikawa H, Yano M, Wakasa K (2014) Genome-wide DNA polymorphisms in seven rice cultivars of temperate and tropical japonica groups. PLoS One 21:e86312
Buntjer JB (1997) Phylogenetic computer tools (PhylTools), ver. 1.32 for Windows, Laboratory of Plant Breeding, Wageningen Univ., Wageningen
Chen K, Wallis JW, McLellan MD, Larson DE, Kalicki JM et al (2009) BreakDancer: an algorithm for high-resolution mapping of genomic structural variation. Nat Methods 6:677–681
Cingolani P, Platts A, le Wang L, Coon M, Nguyen T et al (2012) A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly 6:80–92
Dilday R (1990) Contribution of ancestral lines in the development of new cultivars of rice. Crop Sci 30:905–911
Ebana K, Kojima Y, Fukuoka S, Nagamine T, Kawase M (2008) Development of mini core collection of Japanese rice landrace. Breed Sci 58:281–291
Felsenstein J (1993) PHYLIP (Phylogeny Inference Package) Ver. 3.66. Dept of Genetics, Univ. of Washington, Seattle
Fu Y, Peterson GW, Scoles G, Rossnagel B, Schoen D, Richards K (2003) Allelic diversity changes in 96 Canadian oat cultivars released from 1886 to 2001. Crop Sci 43:1989–1995
Fujino K, Iwata N (2011) Selection for low-temperature germinability on the short arm of chromosome 3 in rice cultivars adapted to Hokkaido, Japan. Theor Appl Genet 123:1089–1097
Fujino K, Sekiguchi H (2005a) Identification of QTLs conferring genetic variation for heading date among rice varieties at the northern-limit of rice cultivation. Breed Sci 55:141–146
Fujino K, Sekiguchi H (2005b) Mapping of QTLs conferring extremely early heading in rice (Oryza sativa L.). Theor Appl Genet 111:393–398
Fujino K, Sekiguchi H (2008) Mapping of quantitative trait loci controlling heading date among rice cultivars in the northernmost region of Japan. Breed Sci 58:367–373
Fujino K, Sekiguchi H (2011) Origins of functional nucleotide polymorphisms in a major quantitative locus, qLTG3-1, controlling low-temperature germinability in rice. Plant Mol Biol 75:1–10
Fujino K, Sekiguchi H, Sato T, Kiuchi H, Nonoue Y et al (2004) Mapping of quantitative trait loci controlling low-temperature germinability in rice (Oryza sativa L.). Theor Appl Genet 108:794–799
Fujino K, Sekiguchi H, Kiguchi T (2005) Identification of an active transposon in intact rice plants. Mol Genet Genomics 273:150–157
Fujino K, Sekiguchi H, Matsuda Y, Sugimoto K, Ono K, Yano M (2008) Molecular identification of a major quantitative trait locus, qLTG3-1, controlling low-temperature germinability in rice. Proc Natl Acad Sci USA 105:12623–12628
Fujino K, Yamanouchi U, Yano M (2013) Roles of Hd5 gene controlling heading date for adaptation to the northern limits of rice cultivation. Theor Appl Genet 126:611–618
Günther T, Schmid KJ (2010) Deleterious amino acid polymorphisms in Arabidopsis thaliana and rice. Theor Appl Genet 121:157–168
Huang X, Kurata N, Wei X, Wang ZX, Wang A et al (2012) A map of rice genome variation reveals the origin of cultivated rice. Nature 490:497–501
International Rice Genome Sequencing Project (2005) The map-based sequence of the rice genome. Nature 436:793–800
Kim H, Hurwitz B, Yu Y, Collura K, Gill N et al (2008) Construction, alignment and analysis of twelve framework physical maps that represent the ten genome types of the genus Oryza. Genome Biol 9:R45
Kim SL, Choi M, Jung KH, An G (2013) Analysis of the early-flowering mechanisms and generation of T-DNA tagging lines in KITAAKE, a model rice cultivar. J Exp Bot 64:4169–4182
Kojima Y, Ebana K, Fukuoka S, Nagamine T, Kawase M (2005) Development of an RFLP-based rice diversity research set of germplasm. Breed Sci 55:431–440
Le Clerc V, Bazante F, Baril C, Guiard J, Zhang D (2005) Assessing temporal changes in genetic diversity of maize varieties using microsatellite markers. Theor Appl Genet 110:294–302
Li R, Yu C, Li Y, Lam TW, Yiu SM, Kristiansen K, Wang J (2009) SOAP2: an improved ultrafast tool for short read alignment. Bioinformatics 25:1966–1967
Murray MG, Thompson WF (1980) Rapid isolation of high molecular weight plant DNA. Nucl Acid Res 8:4321–4325
Nagasaki H, Ebana K, Shibaya T, Yonemaru J, Yano M (2010) Core single-nucleotide polymorphisms-a tool for genetic analysis of the Japanese rice population. Breed Sci 60:648–655
Nei M, Li WH (1979) Mathematical model for studying genetic variation in terms of restriction endonucleases. Proc Natl Acad Sci USA 76:5269–5273
Nonoue Y, Fujino K, Hirayama Y, Yamanouchi U, Lin SY, Yano M (2008) Detection of quantitative trait loci controlling extremely early heading in rice. Theor Appl Genet 116:715–722
Nordborg M, Hu TT, Ishino Y, Jhaveri J, Toomajian C et al (2005) The pattern of polymorphism in Arabidopsis thaliana. PLoS Biol 3:e196
Perrière G, Gouy M (1996) WWW-Query: An on-line retrieval system for biological sequence banks. Biochimie 78:364–369
Roussel V, Leisova L, Exbrayat F, Stehno Z, Balfourier F (2005) SSR allelic diversity changes in 480 European bread wheat varieties released from 1840 to 2000. Theor Appl Genet 111:162–170
Schmid KJ, Sorensen TR, Stracke R, Torjek O, Altmann T, Mitchell-Olds T, Weisshaar B (2003) Large-scale identification and analysis of genome-wide single-nucleotide polymorphisms for mapping in Arabidopsis thaliana. Genome Res 13:1250–1257
Shinada H, Yamamoto T, Yamamoto E, Hori K, Yonemaru J, Matsuba S, Fujino K (2014) Historical changes in population structure during rice breeding programs in the northern limits of rice cultivation. Theor Appl Genet 127:995–1004
Yamamoto T, Nagasaki H, Yonemaru J, Ebana K, Nakajima M, Shibaya T, Yano M (2010) Fine definition of the pedigree haplotypes of closely related rice cultivars by means of genome-wide discovery of single-nucleotide polymorphisms. BMC Genom 11:267
Zhu Q, Ge S (2005) Phylogenetic relationships among A-genome species of the genus Oryza revealed by intron sequences of four nuclear genes. New Phytol 167:249–265
Acknowledgments
This work was supported in part by a grant from the Ministry of Agriculture, Forestry and Fisheries of Japan (Science and Technology Research Promotion Program for Agriculture, Forestry, Fisheries and Food Industry).
Conflict of interest
The authors declare that they have no conflict of interest.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by M. Thomson.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Fujino, K., Obara, M., Ikegaya, T. et al. Genetic shift in local rice populations during rice breeding programs in the northern limit of rice cultivation in the world. Theor Appl Genet 128, 1739–1746 (2015). https://doi.org/10.1007/s00122-015-2543-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00122-015-2543-8