Abstract
Scientific understanding of the genetic components of aging has increased in recent years, with several genes being identified as playing roles in the aging process and, potentially, longevity. In particular, genes encoding components of the nuclear lamina in eukaryotes have been increasingly well characterized, owing in part to their clinical significance in age-related diseases. This review focuses on one such gene, which encodes lamin A, a key component of the nuclear lamina. Genetic variation in this gene can give rise to lethal, early-onset diseases known as laminopathies. Here, we analyze the literature and conduct computational analyses of lamin A signaling and intracellular interactions in order to examine potential mechanisms for altering or slowing down aberrant Lamin A expression and/or for restoring the ratio of normal to aberrant lamin A. The ultimate goal of such studies is to ameliorate or combat laminopathies and related diseases of aging, and we provide a discussion of current approaches in this review.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The nuclear lamina is an array of intermediate filament proteins inside the nucleus of eukaryotic cells, which supports the structure of the nucleus, including its shape and mechanical stability [1, 2]. In addition, it serves as a scaffold for the attachment of DNA–protein complexes that regulate both eu- and heterochromatin histone modifications [3]. The nuclear lamina is involved in the regulation of many key biological processes, including DNA replication, transcription, cell cycle progression, and chromatin organization [2]. Given the central role of the nuclear lamina in such a wide range of essential processes, it is not surprising that alterations in the structure can have a significant impact on normal cellular function, and in some cases can give rise to disease and even mortality within affected organisms.
Maintenance of the nuclear lamina is essential for most eukaryotic life forms, and requires the presence of an array of specific proteins that are highly conserved evolutionarily, both in terms of their structure and function. In particular, major functional components of the nuclear lamina are fibrous proteins known as nuclear lamins, which support this structure through interactions with specific membrane-associated proteins. Lamins are highly conserved evolutionarily, being represented in all examined metazoan life forms; thus, their essential functions likely ensure survival across a broad range of species [4]. Mutations within lamin genes and subsequent alterations in the structure and function of the proteins they encode can give rise to a broad range of diseases known as laminopathies. Such diseases are characterized by a broad range of severe clinical symptoms and complications, with some causing mortality early in life. Numerous laminopathies have been identified in humans during the last decade, and have been linked to several types of mutations in causative loci, both within lamin genes themselves and in genes encoding lamin-binding proteins. Laminopathies include Emery–Dreifuss muscular dystrophy (MIM 181350), dilated cardiomyopathy (MIM 115200), familial partial lipodystrophy (MIM 151660), Charcot–Marie–Tooth disorder type 2B1 (MIM 605588), Greenberg skeletal dysplasia (MIM 215140), limb girdle muscular dystrophy Type 1B (MIM 159001) and mandibuloacral dysplasia with type A lipodystrophy (MIM 248370). The molecular mechanisms by which lamins contribute to these diseases have become increasingly understood in recent years, particularly in terms of the genetic mutations and effects therein on both gene expression and protein structure and function.
Lamin A processing, mutations, and role in diseases
Lamins can be categorized as either A type (lamins A and C) or B type (lamins B1 and B2). In humans, A-type lamins are encoded by a single gene—LMNA (Entrez Gene ID: 4000)—located on chromosome 1q21.2, while B-type lamins are encoded by two genes—LMNB1 and LMNB2 (Entrez Gene ID: 4001 and 84823)—located on chromosomes 5q23.2 and 19p13.3, respectively. The processes involved in the expression of lamin genes and their translation and processing into mature and functional proteins include a series of specific and essential steps, alterations to which can impact the essential molecular and cellular functions of these proteins. In the case of LMNA, one essential step in protein biosynthesis and maturation is farnesylation at the C-terminus by the enzyme farnesyltransferase [5]. This posttranslational modification plays a role in targeting prelamin A to the inner nuclear membrane. Farnesylation is followed by several steps involving the endoproteolytic cleavage of the last three amino acids by zinc metallopeptidase ZMPSTE24, carboxymethylation of the C-terminal cysteine by ICMT methyltransferase, and proteolytic removal of the last 18 amino acids by ZMPSTE24, resulting in the removal of the farnesyl tail on the C-terminus [6, 7]. Mature lamin A is then released from its membrane anchor, which allows it to be properly positioned in the nuclear scaffold. Factors that interfere with these steps in such a way as to affect lamin maturation can have negative effects on nuclear lamin and can ultimately lead to an array of downstream effects, detrimental to cellular health and in some cases, longevity.
Within the LMNA gene alone, over 400 different point mutations have been identified, many of which are underlying causes of laminopathies [8, 9], including restrictive dermopathy (MIM 275210) and Hutchinson–Gilford progeria syndrome (HGPS; MIM 176670). HGPS presents as a broad range of clinical features, which most notably include accelerated aging [10]. HGPS is caused by mutations in the LMNA gene, the most well-known of which is a de novo heterozygous point mutation in position 1824C > T (G608G) [11]. While the G608G mutation does not cause any change in the encoded amino acid, it does activate a cryptic splice donor site in exon 11 of the LMNA gene. Consequently, a splice variant of Prelamin A mRNA is generated with an internal deletion of 150 base pairs [11]. These transcripts are translated into progerin, the truncated form of the lamin A protein, with a 50 amino acid internal deletion near the C-terminus [11]. The internal deletion eliminates the essential endoprotease ZMPSTE24 recognition site, resulting in progerin remaining permanently farnesylated and anchored to the nuclear membrane [11, 12]. The accumulation of progerin in cells of patients carrying the G608G mutation severely impacts the structure of the nuclear lamina, culminating in the cellular and disease phenotypes characteristic of HGPS.
Severe forms of progeria also occur due to a number of other mutations in LMNA, such as 1821 G > A and 1,968 G > A, mutations associated with increased ratios of progerin to normal, wild-type protein [13]. An extremely severe case of neonatal progeria in which death occurs within the first year of life has recently been found to be associated with heterozygosity (1,821 G > A). Examination of patient fibroblasts demonstrates an increased ratio of progerin to lamin A, relative to those levels typically observed in HGPS, suggesting that disease severity may be determined in part by the ratio of the farnesylated protein to mature lamin A [13, 14].
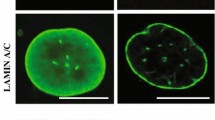
The hallmarks of progeria and its characteristic phenotypes are broadly associated with alterations in the production of progerin relative to mature lamin A, imbalances that directly impact key biological processes occurring at both the genetic and cellular levels. Progerin is observed to accumulate in all tissues of HGPS patients, acting as a dominant-negative protein that significantly modifies the structure of the nuclear lamina [15]. The cellular phenotype of HGPS patients includes nuclear blebbing, thinning of the nuclear lamina, loss of peripheral heterochromatin, and clustering of nuclear pores [16]. Accumulation of progerin in HGPS as nucleoplasmic aggregates leads to inhibition of the transport of several factors that play key roles in the functioning of the nucleus [15]. Examination of fibroblast cells from patients with HGPS demonstrates deficiencies in histone modification, alterations in gene expression, delays in the response to DNA damage, disturbances of mitosis, and cytokinesis, abnormalities in chromosome segregation and increases in the occurrence of binucleated cells [9]. The hallmarks and clinical features of HGPS are therefore deeply rooted in alterations taking place at genetic and protein biosynthesis levels and, in turn, those subsequent changes that negatively impact key biological processes further downstream. While there are significant components to HGPS that are associated with aging, disease pathogenesis, and progression is likely to involve several factors not exclusive to the aging process. While many tissues in HGPS patients exhibit phenotypes associated with accelerated aging, not all tissues are typically affected (reviewed by [17]). In addition, HGPS may not be viewed exclusively as a disease of accelerated aging, given that certain aspects of the disease are not typically associated with normal aging; for example, the presence of clavicular agenesis. Significant features of HGPS that are associated with normal aging include increases in DNA damage, defects in DNA repair, alterations in telomeric dynamics, and increases in cell proliferation, senescence, and tissue homeostasis (reviewed by [17] and references therein). In this respect, HGPS may be viewed as a disease that substantially resembles premature aging, but does not include all aspects of it, and is segmental in nature.
The significance of LMNA in human health and longevity
Several genes have been identified in recent years as influencing the aging process and possibly longevity [18, 19]. The potential significance of the LMNA gene in human health and its potential contribution to susceptibility to many common diseases is also becoming increasingly appreciated. In particular, Scaffidi et al. demonstrate that the molecular mechanism that underlies HGPS also takes place in normal cells at a lower rate [20]. The nuclei of cells of normal-aged individuals exhibit defects similar to those of cells of HGPS patients, including changes in histone modification and increased levels of DNA damage. Age-dependent defects in the nuclei of cells of healthy individuals are caused by infrequent use of the same cryptic splice site of Lamin A, whose constitutive activation generates a Progerin transcript [20]. The over-expression of normal Prelamin A can lead to growth defects in human vascular smooth muscle cells [21], similar to those changes observed in cells producing Progerin [22]. Cytotoxicity can also be induced by a minor increase in the steady-state level of one or more intermediate products of Prelamin A processing [12, 22].
Recent years have shown extensive investigation of the potential contribution of genetic variability within lamin genes to disease susceptibility. Disease-association studies including SNPs at lamin loci, have implicated metabolic syndrome, dislipidemia, type-II diabetes, obesity, polycystic ovary syndrome, arterial stiffness, and vascular disease [23–35]. In addition, there is some evidence for the potential influence of genetic variation at LMNA on human longevity and age-related diseases [36–38]. Findings from these studies have been variable, with the majority focusing on the 1908C > T; rs4641 LMNA SNP. rs4641 has been found in several cases to be significantly associated with disease susceptibility and related conditions across a number of ethnically diverse population cohorts for type II diabetes and related diseases [23, 24, 26–28, 31]. The rs4641 SNP is a silent C > T substitution occurring at exon 10 of the LMNA gene, the exon in which alternative splicing gives rise to mRNAs that code for either Prelamin A or Lamin C [39]. The mechanism by which this SNP alters the LMNA gene product and phenotype to potentially influence susceptibility to these diseases is unknown. However, recent evidence suggests that the C and T alleles of rs4641 are associated with differential gene expression phenotypes, with the C allele associated with increased levels of transcripts of Lamin A and Lamin C relative to those detected for the T allele [40]. While this study demonstrates that differential, allele-specific expression is present at the LMNA locus in HGPS, it is unclear whether or not such variability is directly associated with the rs4641 SNP or if it is rather associated with other variants located within the same haplotype block. In light of these studies, the relevance of genetic variation at the LMNA locus to more common diseases affecting populations at large may be significant. Interestingly, the rs4641 SNP is represented in all populations that have been examined in the HapMap project to date, with the minor ‘T’ allele represented at levels ranging from between 5 and 10 % in African populations, 20–25 % in Europeans and 23–32 % in South and East Asian populations (www.hapmap.org) [41]. Elucidating the role of this relatively common SNP in disease pathogenesis, longevity, or related diseases, therefore, may have broad significance. However, in-depth examination of linkage disequilibrium between rs4641 and other functional SNPs is required to delineate the role of this LMNA SNP in human diseases, metabolic-related, age-related, or otherwise.
Given the evidence that genetic variation at LMNA contributes to both laminopathies and more common human diseases, the identification of methods that can therapeutically alter LMNA structure and restore a healthy homeostatic balance of aberrant/normal LMNA, warrants further investigation. A multifaceted approach is required to increase knowledge in this area and further elucidate the functional relevance and complex characteristics of lamins both in terms of their expression and functional interactions. In this way, interventions may be designed and developed to intervene, treat, and ameliorate symptoms of human diseases, particularly those associated with aging.
Targeting LMNA and associated diseases: a multifaceted approach
Combating the broad range of consequences associated with missplicing lamins and altering their gene expression levels requires a multifaceted approach, which includes targeting components at the genetic level as well as targeting components downstream cell signaling and cellular-level processes. Furthermore, unraveling the underlying complexity at each of these levels and, in turn, targeting specific processes therein to treat or ameliorate disease symptoms, requires (a) an in-depth knowledge of the molecular interactions between nuclear lamins and other proteins and cellular events, (b) in vitro and in vivo studies demonstrating effectiveness of the treatment and validating such interventions, and (c) development and refinement of techniques to manage, limit, and potentially reverse damage that has already been incurred in patients. Here, we perform an analysis of the existing literature and published data sets, with the goal of identifying novel targets for treating laminopathies and associated diseases. In turn, we hope that these findings may provide a basis for future experimental design, interpretation of results, and refinement of methods aimed at tackling severe laminopathies and other age-related diseases.
Computational analysis of LMNA signaling and intracellular interactions
In this study, we utilized literature searches of the NIH’s PubMed database in order to examine pathways that regulate LMNA expression. This was accomplished by using the following keywords during searches: gene expression regulation Lamin A/C or Progerin, and progeria. In order to visualize the molecular interactions between nuclear lamins and other proteins and identify novel targets, the Ingenuity Pathway Analysis (IPA, Ingenuity© Systems, www.ingenuity.com) software and knowledgebase and its pathway designer graphical module were utilized. IPA provided graphical representations of network interactions of the LMNA protein with molecules involved in signal transduction pathways and other intracellular regulatory networks. Data used to generate pathways and interaction networks in IPA are compiled from interactions validated in multiple model organisms from peer-reviewed journals by a team of IPA scientists. Advantages of this software tool include: (a) each connection displayed on a graph is documented by a peer-reviewed article, which can be examined by clicking on the relevant connection and (b) the Pathway Designer module contains “cell art” elements which can be used to graphically display connection locations (nucleus, mitochondria, cellular membrane, etc.).
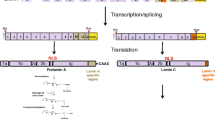
Searches did not identify signaling or metabolic pathways in IPA that center on LMNA. However, LMNA was found to be part of a canonical Apoptosis Signaling pathway as a target of caspase 6. The LMNA Interactome generated through this analysis displays 110 direct molecular interactions with LMNA, with all the known molecules that LMNA/lamin A interacts with, including proteins, protein modifiers, small molecules, and microRNAs. Some of these molecules are members of other signal transduction pathways, and therefore represent a bridge between LMNA and these pathways. The most important signal transduction pathways that target/affect LMNA are shown in Fig. 1. Data shown in this figure was generated by combining information from several individual canonical pathways, the LMNA Interactome, and additional information from the published literature, and then repeating several iterations of this process to reach a final model. As shown in Fig. 1, WNT/beta-catenin, TGF beta, Notch, and PI3K represent the key signaling pathways upstream of LMNA, which likely regulate its expression. The main molecules that interact with the lamin A protein, and have genetic correlates with some of the laminopathies we have discussed here, are mainly found in the nucleus, including Sun1 and Sun2, whose potential roles in HGPS disorder have been investigated recently [42]. Many of the identified signaling pathways and molecules that interact with lamin A are known to exert effects on nuclear lamins by altering their expression levels. To visualize gene expression regulatory points that potentially may be targeted for intervention, the Pathway Designer module was applied to the data presented in Fig. 1. Potential interventions might include following mechanisms: transcription, splicing, translation, posttranslational modification, and degradation via autophagy. Using the overlay function of IPA, this figure was overlaid with a number of pharmaceuticals and drugs that may be used to target key proteins of any given biological process. While a number of drugs were identified via the IPA database, more detailed lists of drugs and agents that may be applicable are listed in Tables 1, 2, 3, 4 and 5 generated through PubMed searches using relevant keywords. To expand the literature search and identify additional known drugs and experimental compounds, as well as their side effects, potentially acting on elements of the pathways involving LMNA gene, we employed a manually curated proprietary database (MetaCore™, GeneGo), and the MetaCore pathway analysis software.
Key signaling pathways upstream of LMNA, which regulate its expression (listed mainly on the top of the figures), and the main molecules that interact with lamin A/C protein (listed mainly in the nucleus). Labels denoting physiological processes in the cell are derived partially from the IPA listing under category “Top Functions and Diseases”: Mechanical Stability of the Nucleus, Response to DNA Damage, Gene Transcription, Cell Cycle Progression. The meanings of the molecular symbols are described in the figure legend, which is part of the figure. Lines represent interactions between the molecules. They can have arrows, solid lines or nothing at the end, which represent directional action, inhibitory action, or just binding of two molecules, respectively. Red crossing lines in the nucleus represent a DNA double helix. Entrez Protein Names and their symbols used in figures, are in parenthesis: Catenin (cadherin-associated protein), beta 1, 88 kDa (β-catenin), beta-transducin repeat containing (β-TrCP), eukaryotic translation initiation factor 4E binding protein 1 (eIF4EBP), AHR ligand, aromatic hydrocarbon (AHR Ligand), protein kinase B/Akt (AKT), Adenomatous polyposis coli, APC (APC), mitogen-activated protein kinase kinase kinase 5 (ASK1), adenosine 5′-triphosphate (ATP), axin 1 (Axin), barrier to autointegration factor 1 (BANF1), BCL2-associated X protein (BAX), B cell CLL/lymphoma 2 (Bcl-2), beta-catenin-LEF/TCF (Betacatenin/TCF), V-raf-1 murine leukemia viral oncogene homolog 1(c-Raf), cadherin (E, N, P, VE), calcium-dependent protease, M calpain (Calpain), CASPASE-1 (Caspase), interleukin 1 converting enzyme (ICE), caspase 12 apoptosis-related cysteine peptidases (Caspases 2,3,6 and 12), casein kinase I (CKI), cyclic AMP (cAMP), eukaryotic translation initiation factor 2-alpha kinase 2 (EIF2AK2), eukaryotic translation initiation factors (eIF 4A, 4B, 4G, and 4E), emerin (EMD), p42/p44 MAP kinase (ERK1/2), filamentous actin (F Actin), PTK2 protein tyrosine kinase 2 (FAK), FBJ murine osteosarcoma viral oncogene homolog (FOS), frizzled (FZ, FZD), MTOR-associated protein, LST8 homolog (S. cerevisiae) (GBL), frequently rearranged in advanced T cell lymphomas (GBP), glucagon receptor (GCGR), growth hormone receptor (GHR), growth factor receptor-bound protein 2 (GRB2), growth hormone (GH), glycogen synthase kinase 3 (GSK3), hydrogen peroxide (H 2 O 2 ), HtrA serine peptidase 2 (HtrA2), I KAPPA B (IκB), insulin-like growth factor 1 (somatomedin C) (IGF-1), insulin-like growth factor 1 receptor (IGF1R), I kappa β-NF-kappa β (Iκβ-NFκβ), integrin-linked kinase (ILK), mitogen-activated protein kinase 8 (JNK1), Jun proto-oncogene (JUN) lamin A/C (LMNA), LEM domain containing 3 (LEMD3), myelin-associated glycoprotein (MAG), p53 binding protein homolog (mouse) (MDM2), methyl CpG-binding protein 2 (Rett syndrome) (MECP2), MAP kinases (MEK1/2, MKK4/7, MKK4/7), mechanistic target of rapamycin (serine/threonine kinase) mTORC1 (mTOR), V-myc myelocytomatosis viral oncogene homolog (avian) (MYC), NF-KAPPA B(NF-kB), cyclin-dependent kinase inhibitor 2A (melanoma, p16, inhibits CDK4) (p14ARF), tumor protein p53 (p53, TP53), microtubule affinity-regulating kinase 2 (MAP), proliferating cell nuclear antigen (PCNA), 1-phosphatidylinositol 3-kinase (PI3K), peptidylprolyl cis/trans isomerase, NIMA-interacting 1 (PIN1), protein kinase C (PKC), phospholipase C gamma, (PLCG), protein phosphatase type2a (PP2A), peroxisome proliferator-activated receptor gamma (PPARG), protein kinase C, alpha (PRKCA), parathyroid hormone (PTH), parathyroid hormone 1 receptor (PTH1R), regulatory associated protein of MTOR, complex 1″ (Raptor), p21 Ras (Ras), retinoblastoma 1 (RB1), Ras homolog enriched in brain (Rheb), (Src homology 2 domain containing)-transforming protein 1 (SHC), SMAD family members (Smad3 and Smad4), V-src sarcoma (Schmidt-Ruppin A-2) viral oncogene homolog (avian) (Src), sterol regulatory element-binding transcription factor 1 (SREBF1), signal transducer and activator of transcription 4 (STAT4), Sad1 and UNC84 domain containing 1 (SUN1), Sad1 and UNC84 domain containing 2 (SUN2), spectrin repeat containing, nuclear envelope 1 (SYNE1), spectrin repeat containing, nuclear envelope 2 (SYNE2), TGF beta (Tgfβ), thyroid hormone (T3), thyroid hormone receptor (TR), polyubiquitin (Ub), von Hippel–Lindau tumor suppressor (VHL), WNT inhibitory factor 1 (WIF1)
Targeting Lamin A and Progerin expression via signal transduction pathways
On the basis of the computational analysis outlined above, a number of potential targets and therapeutic interventions have been identified and discussed. Many of these potential interventions may nonspecifically down-regulate the level of expression of both Lamin A and its disease-associated allelic variants, while others have more specific effects on mutant Lamin A expression.
Restoration of IGF-1 and GH balance
Insulin-like growth factor 1 (IGF-1) signaling is involved in aging and longevity in many animals, including nematodes, Drosophila, and mammals (for review see [43]). Zmpste24 (−/−) mice, a mouse model of progeria, exhibits dysregulation of somatotropic axis, characterized by high levels of circulating growth hormone (GH) and reductions in insulin-like growth factor-1 (IGF-1) [44]. Application of recombinant IGF-1 restores the balance between IGF-1 and GH, and this delays the onset of several progeroid characters and prolongs the lifespan of progeroid animals [44, 45]. However, applying such an approach as a means of treating HGPS may be limited, given the diverse biological effects exerted by this hormone, and in particular, the pathogenic role of IGF-1 signaling in cancer [46].
Notch signaling inhibitors
Expression of Progerin ectopically activates effectors of Notch and downregulates the canonical Wnt signaling pathway, regulating the differentiation of mesenchymal stem cells [47, 48]. This leads to misregulation of somatic stem cell differentiation, explaining some of the pathological defects of HGPS [49]. Thus, inhibitors of Notch signaling (Table 1) and recombinant β-catenin could potentially ameliorate symptoms of HGPS. Notch 2 may also be targeted for inhibition given its influence on Granzyme B transcription [50] and potentially apoptosis; however, the impact of such an approach on immune function is unclear. Furthermore, there are severe side effects known to be associated with certain Notch inhibitors, including gastrointestinal bleeding and skin cancer [51]. Therefore, caution must be taken when applying such approaches to treating HGPS patients.
Reactive oxygen species scavengers
Basal levels of reactive oxygen species (ROS) as well as induced levels of H2O2 are five times higher in HGPS fibroblasts compared to normal fibroblasts, which leads to double stranded breaks (DSBs) in DNA and a decrease in the proliferative capacity of cells [52]. Indeed, HGPS is accompanied by an elevated quantity of DSBs and attenuation of their repair [53]. On the contrary, the ROS scavenger N-acetyl cysteine (NAC) has been shown to decrease basal levels of DSBs and enhance population-doubling times in fibroblasts derived from HGPS patients [52]. Other effective ROS scavengers are listed in Table 2. However, the effectiveness of using ROS scavengers for treating progeria patients and ameliorating intracellular damage and associated symptoms remains somewhat speculative. In addition, the complex biochemical effects of some anti-oxidants may raise some safety concerns. While the anti-oxidative effects of ascorbic acid are well characterized, pro-oxidative effects have also been described [54–56] and such properties must be taken into consideration when developing treatments for progeria.
Telomerase activators
Progressive attrition of telomeres causes activation of progerin production in normal human fibroblasts [9]. Active telomerase prolongs the cellular lifespan of HGPS by decreasing progerin-induced DNA-damage signaling and activation of both the p53 and Rb pathways. These are the two pathways that mediate the onset of premature senescence in HGPS [57]. Telomerases can be stimulated by a potent telomerase activator, TA-65, to extend short telomeres, and this has been shown to have a positive effect on health span in mice [58]. Treatment with TA-65 may prevent some symptoms of HGPS. However, while the results in mice are promising, the use of this technology in humans may be limited, particularly in the event of any significant side effects being identified in future clinical trials.
Rb regulators
Progerin accumulation leads to premature replicative cellular senescence [59]. Marji et al. suggest, based on global gene profiling of HGPS fibroblasts, that defects in the lamin A-Rb signaling pathway may be key factors in the accelerated aging phenotype of HGPS, and perhaps in normal aging, too [60]. Rb activity can be modified with reagents such as roscovitine and PD-0332991, inhibitors of Cdk2–cyclin E and Cdk4/cyclin D1 complexes, respectively, that phosphorylate and inactivate the Rb tumor suppressor [61]. However, studies indicate that Rb expression is decreased in fibroblasts in both HGPS and normal aging, with a concomitant reduction in phosphorylation [60, 62]. In this respect, an intervention that increases Rb expression and/or increases Rb phosphorylation to normal physiological levels may provide some therapeutic benefit in HGPS.
Apoptosis inhibitors
Nuclear progerin accumulation leads to accelerated aging and increased apoptosis in individuals suffering from HGPS [59], and the same observation has been made in aging HGPS fibroblasts [63]. Theoretically, apoptosis inhibition (Table 3) may offer some means of extending the cellular lifespan of HGPS patients. However, the potential for increased risk of developing cancers and other related diseases would almost certainly limit such an approach.
Inhibitors of translation and autophagy activators
Rapamycin, an immunosuppressant drug, delays cellular senescence and organismal aging, abrogates nuclear blebbing, and stimulates degradation of progerin in HGPS cells [64, 65]. This drug can selectively decrease progerin levels in progeria cells through a mechanism involving autophagic degradation [66]. Rapamycin treatment decreases the formation of nonsoluble aggregates of progerin and induces progerin elimination by autophagy in normal fibroblasts [64]. A safer alternative to rapamycin, rilmenidine, a centrally acting anti-hypertensive drug, was found to induce autophagy in cell culture via a pathway independent of the mammalian target of rapamycin [67]. As a natural alternative to the acid form of tretinoin (all-trans-retinoic acid), vitamin A has been found to induce autophagy. The essential oil produced from rose hip seeds is a natural source of tretinoin and promotes autophagosome maturation through a pathway independent from the classic nuclear hormone receptors [68, 69]. Another natural autophagy activator is vitamin K2 [70]. A comprehensive list of known autophagy activators is listed in Table 4.
Several small molecules down-regulate lamin A/C protein via mechanisms of proteolysis. Doxorubicin (also known as adriamycin) is a topoisomerase II inhibitor used in anti-cancer therapy whereby it induces activation of caspases, leading to cleavage of lamin A/C [71]. Doxorubicin has also been identified in a high-content screen for inducers of autophagy [72]. Sangivamycin is a nucleoside analog that acts via activation of JNK and protein kinase C delta. In MCF-7/Adr cells, sangivamycin increases cleavage of human lamin A/C protein [73]. Tunicamycin and thapsigargin, endoplasmatic reticulum (ER) stress inducers, increase degradation of mouse LMNA protein via the activation of caspases [74]. Paclitaxel, a chemotherapeutic agent, induces cleavage of lamin A/C, enhanced by the synthetic peptides Smac/DIABLO [75]. It should be noted that HGPS is characterized by a large increase in the rate of apoptosis [63], and while application of apoptosis inducers in HGPS treatment is restricted, autophagy inducers can be considered safe, since they do not induce cell death [72].
cAMP activators
Several hormones have been identified as having effects on the expression, function, or phosphorylation of Lamin A. Hormones that increase cAMP levels (glucagon, calcitonin, vasopressin, and parathyroid hormone (PTH)) decrease lamin A protein phosphorylation of rat LMNA protein in renal medullary thick ascending limb cells [76]. cAMP-dependent phosphorylation controls nuclear lamin associations, and aberrant phosphorylation could cause remodeling of the lamina [77]. Therefore, hormone modulation of lamin A phosphorylation with glucagon, calcitonin, vasopressin, or parathyroid hormone might be another way to alleviate laminopathy symptoms.
Thyroid hormone supplementation
Thyroid hormone (T3) decreases expression of mouse Lamin A mRNA in liver from mice exhibiting hypothyroidism [78]. An association has been reported between low levels of T3 and DeBarsi syndrome, an autosomal recessive syndrome characterized by a progeria-like appearance [79, 80]. The endocrine system is affected by aging, and while T3 has been associated with longevity, deficiencies in, or suboptimal levels of T3 are more common in older individuals, particularly women. Therefore, supplementation with T3 might have beneficial effects on progeria symptoms, as well as aging [81, 82].
PI3K pathway inhibitors
In T98G cells stimulated with the growth factor PDGF, inhibition with a small molecule inhibitor of PI3K (LY294002), which prevents Akt phosphorylation, has been found to decrease expression of human LMNA mRNA induced by the PDGF-BB. This demonstrates that LMNA may be regulated through the phosphatidylinositol 3-kinase PI3K pathway [83]. It has been suggested that aberrant phosphorylation of Ser458 of Lamin A by Akt1 contributes to striated muscle laminopathies caused by LMNA mutation [84]. Therefore, inhibition of Akt1 by LY294002 might be beneficial to this and similar laminopathies.
Epigenetics marks reversal
Reversal of epigenetic marks may represent a novel anti-aging target. In an animal model of progeria, Zmpste24-deficient mice show hypermethylation and transcriptional silencing of rDNA genes. This effect is reversible through treatment with methyltransferase inhibitors [85]. Therefore, methyltransferase inhibitors (Table 5) could prevent HGPS symptoms. In the same animal model it has been noted that a delayed DNA damage response is a result of histone H4 acetylation defect. Reversal of this defect by supplying the histone deacetylase inhibitor sodium butyrate in drinking water ameliorated aging-associated effects, and extended the lifespan in the animal model. In addition to accumulation of progerin, aged mice show hypoacetylation of the histone H4K16 [86]. Therefore, treatment with methyltransferase inhibitors (Table 5) and histone deacetylase inhibitors could potentially reduce HGPS symptoms. However, such interventions could give rise to significant side effects and would have to be carefully evaluated and refined before transfer to the clinical setting.
Targeting posttranslational modification: farnesylation inhibitors
Ionafarnib (SCH-66336), a farnesyltransferase inhibitor, has been shown to inhibit prelamin A farnesylation in buccal mucosa cells [87]. Other studies demonstrate that inhibitors of farnesyltransferase (FTIs) ameliorate the phenotype of transgenic G608G LMNA mice [88]. This model is characterized by extensive and progressive loss of vascular smooth muscle cells (VSMCs) of large arterial media [89], similar to effects observed in human HGPS patients [90, 91]. FTIs have also been shown to improve survival and bone integrity in LMNA HG/+ [92, 93] and in ZMPSTE24−/− mouse models [94]. The compound FTI-277 may completely restore localization of nuclear antigens in HGPS fibroblasts [59]. The combination of statins and aminobisphosphonates has been shown to inhibit the production of farnesylation and geranylgeranylation modifications of prelamin A and progerin in Face-1/Zmpste24-defective mice, decreasing senescence-like symptoms and increasing the lifespan of affected mice [95, 96]. Unfortunately, the FTI treatment has harmful side effects such as centrosome separation and bipolar spindle formation defects, nuclear dysmorphy, and cytotoxicity [97]. In addition, mice, expressing nonfarnesylated progerin variants (LMNA(nHG/+)), still reveal progeria-like phenotypes, which are not ameliorated by FTI [98].
Directly and selectively targeting mutant Lamin A
Targeting mutant Lamin A RNA: antisense oligonucleotides, RNAi, miRNA, and siRNA
Inhibition of the LMNA miss-spliced site reverses senescence-associated defects in cell nuclei [20]. Fong et al. demonstrated the effectiveness of antisense oligonucleotide technology and identified an antisense oligonucleotide which is complementary to a site in exon 11 at a 5′ position relative to the alternate splice site in LMNA transcripts. This may be used to decrease alternative splicing in HGPS fibroblasts and moderately reduce progerin levels [99]. Splicing-based therapeutical approaches have been examined using a genetically modified mouse strain that carries an HGPS mutation. Antisense morpholino-based therapy has been developed with the aim of preventing pathogenic LMNA splicing, and alleviating the progeroid phenotype [100].
For the past decade, cancer treatments have been developed based on RNA interference (RNAi)—a mechanism that effectively “shuts down” malfunctioning genes with small noncoding RNA molecules from the families of microRNAs. LMNA transcripts have a myriad of microRNAs with which they specifically interact; hence, an RNAi approach offers potential for targeting misspliced LMNA transcripts. In the brain, the Prelamin A transcript is regulated by the brain-specific microRNA miR-9. The tight shutdown of the LMNA transcript observed in the brain using miR-9 may explain the lack of central nervous system pathology in mouse models of HGPS [101]. Recently, Weidefield et al. have reported on the generation of a conditional inducible microRNA (RNAi) system for the controlled inactivation of LMNA [102]. There is also evidence of differential expression of miRNAs in control versus LMNA-related laminopathy [103]. A systemic application of siRNA, specifically targeted to tissues of interest may offer promising potential in future therapeutic applications.
Targeting mutant protein accumulation: chemical protein binding
A proteomics approach using matrix-assisted laser-desorption-ionization time of flight (MALDI-TOF) MS [104] has identified that lamin A belongs to a family of poly (ADP-ribose) binding proteins. Nuclear lamin A was found to be covalently bound to acetaminophen (APAP), and also appears to become phosphorylated upon arylation. Lamin A may be associated with disruption of nuclear membrane organization, which may be triggered by the translocation of the 55- to 58-kDa APAP-protein adduct, leading to cell death [105]. Direct lamin A/chemical binding may be explored by designing a molecular sponge that sequesters mutant lamin A, i.e., progerin from the cell.
Gene therapy: nanotherapy, viral vectors, protocells, and targeting progerin for autophagy
Several gene therapies and nanotherapies targeting cellular proteins are currently in development. One such approach to the aggregation of misfolded proteins has been applied in the case of a Hungtinton’s neurodegeneration (HD) mouse model (HDR6/1) by targeting proteins for autophagic proteosomal degradation using intrabodies. This may represent an effective strategy if modified for clearance of progerin instead [106]. Genome customization and targeted gene modification of Lamin A mutant alleles using gene-specific engineered nucleases such as zinc finger nucleases or transcription activator-like effector nucleases (TALENs, Cellectis) represents another possible approach [107].
The assortment of gene delivery vehicles for gene therapy products are expanding, and include lentivirals and adeno-associated viral vectors [108] in addition to non-viral gene delivery systems such as lipoplexes, polyplexes, inorganic nanoparticles, quantum dots, and protocells [109–111]. However, despite significant progress in recent years, limitations still persist in refining this technology for use in the clinical setting both in terms of patient safety and efficacy.
Future directions
The role of lamin A is to maintain nuclear structure and integrity and in doing so, contribute to the health and survival potential of individuals within a population. The consequences of defectiveness in lamin A structure and function are observed, therefore, to be extremely severe, manifesting at several broad-ranging and essential biological levels, which include cell signaling and gene expression. As such, any intervention successful in ensuring the maintenance of LMNA protein function and/or the reduction of LMNA’s downstream effects must work within key parameters at each of these regulatory levels. Our analysis has identified potential targets for therapeutic intervention by addressing both causes and effects of LMNA defectiveness. Our findings provide a framework for targeting LMNA defectiveness directly at the genetic level and further downstream by targeting signaling events and other processes which give rise to cellular insult and ultimately disease.
By means of a computational analysis of multiple biological pathways, we have identified a number of plausible therapeutic targets, and outlined 12 possible interventions for regulating defective LMNA expression and protein accumulation (see Fig. 2). These are: (1) IGF-1 and GH balance restorers, (2) Notch signaling inhibitors, (3) reactive oxygen species scavengers, (4) telomerase activators, (5) Rb inhibitors, (6) apoptosis inhibitors, (7) translation- and autophagy-activator inhibitors, (8) cAMP activators, (9) thyroid hormone supplementation, (10) PI3K pathway inhibitors, (11) epigenetics marks reversal, and (12) farnesylation inhibitors. While these targets are highly specific in many cases, collectively they are wide ranging and cover the biological complexities that characterize LMNA-related diseases and the levels at which they manifest. Furthermore, we have presented a comprehensive list of compounds known to act on specific targets within these biologic pathways. A combinatory approach may be applied using this data to develop a therapy or therapies consisting of a combination of several key compounds, potentially including: farnesylation inhibitors, autophagy activators, apoptosis inhibitors, and telomerase activators.
Presented are steps of the gene expression cycle of Lamin A/C that can be targeted for additional regulation: transcription, splicing, translation, posttranslational modification, and degradation via autophagy. See text for a description for strategies for targeting each of these steps with already-available drugs to minimize the deleterious effects of altered Lamin A expression in laminopathies, and possibly aging. Gene names and symbols are the same as those listed in Fig. 1. Red dots represent farnesylation residues on lamin A/progerin
Extensive in vitro studies characterizing the effectiveness of these compounds are required before moving towards translation into clinical practice. In particular, the effects of these compounds on progerin accumulation is of particular interest and may be used as a measure of treatment efficacy; in part, it could also be used in the validation of such interventions. However, there are several outstanding questions that must be answered in order to validate the effectiveness of such interventions and the mechanisms by which they provide benefit. Outstanding questions in this field concern the molecular mechanisms by which laminopathies and related diseases manifest and the processes in which progerin alters cellular phenotype and biological age.
In addition to identifying molecular targets, the approach outlined here has also focused on identifying molecules that can carry out specific functions. The use of small molecules to activate/inhibit the signal transduction pathways that regulate LMNA expression itself represents one such approach. In this manner, lamin A in addition to progerin may be down-regulated to levels that effectively influence cellular homeostasis to a point at which cells are healthy, producing lower levels of both laminA and progerin, and therefore potentially lowering the rate at which aging occurs. There are many small molecules/drugs presently available and approved by the FDA (albeit for other purposes), which may be used to target signaling pathways in this way (Tables 1, 2, 3, 4 and 5). However, as part of this strategy, it will be necessary to further characterize the significance of the ratio of progerin/laminA in disease pathogenesis. A long-term goal may be to develop a method of directly targeting of the underlying causes of progeria and related conditions. This requires a technological platform that targets and discriminates progerin and other products of disease-causing Lamin A alleles, from functional lamin A. There are potential tools available at present that may be tailored further for this purpose. In particular, it is feasible that progerin may be specifically targeted at the mRNA level using RNAi tools. In order to target toxicity associated with mutant protein accumulation, designer proteases may also be developed to specifically degrade the mutant protein. In addition, the use of intrabodies to bind and target progerin for autophagy also represents a potential means for achieving these aims. Delivery mechanisms will also be key to any such therapeutic intervention, and the use of viral vectors and nanotherapeutic delivery approaches hold much promise as they are developed and refined into the future. Antisense morpholino-based gene therapy also holds much promise. By directing this technique to prevent pathogenic LMNA splicing, Osorio et al. have achieved reductions in progerin accumulation and associated nuclear defects, amelioration of progeroid phenotypes, and an extension of lifespan [100]. While the gene therapy approach requires further refinement, this technology clearly represents the most likely means of ensuring correct splicing and localization of defective LMNA. Drugs which enhance autophagic mechanisms to achieve reductions in progerin accumulation also show considerable potential; however, safer analogues are required [112]. Another approach may be the use of small molecules to ameliorate the effects of progerin accumulation, such as those listed in this review. However promising, further evaluation of the potential impact of these molecules on the disease phenotype are required prior to application in the clinical setting, either individually or in combination with other therapies. Currently, a number of clinical trials are underway to examine the potential therapeutic effects of using statins, FTIs, and bisphosphonate in combination to treat progeria [113, 114]. Similarly, some of the molecules highlighted in this study may be incorporated into future clinical trials or treatments.
The overall focus of this review has been to identify and highlight different methods that may be used for treating laminopathies, and to a lesser extent, other LMNA-associated human diseases and aging. While some treatments may act to target the downstream effects of progerin accumulation, other treatments may be used to directly alter the ratio of progerin/wild-type protein. Indeed, the most effective method of treating laminopathies would be to target and counteract progerin accumulation directly. However, the targeting of LMNA expression in general also holds potential for treating patients given that disease severity may be determined in part by the ratio of progerin to mature lamin A. In order for future treatments to significantly alter the ratio of wild-type/mutant protein in favor of cellular health and longevity, mechanisms that achieve increments comparable to wild-type protein levels may be required in addition to also reducing progerin levels. Modulating the expression of Lamin A may also be effecive for the treatment to other human diseases associated with LMNA, given that differential allele-specific expression has been identified at the LMNA locus [40]. For example, alleles represented at relatively high frequencies in human populations have been associated with a number of relatively common human diseases (e.g., rs4641). In this respect, development of methods of increasing expression of wild-type LMNA may offer a means of treating both laminopathies and other human diseases association with genetic variation at this locus.
The consequences to intervening to alter the ratio of Progerin/LMNA expression are likely to be significantly influenced by a host of factors, including underlying differences in cell-type and tissues to which treatment is directed. In particular, LMNA expression is known to be developmentally regulated, being expressed in differentiated cells while being absent from early embryonic stem cell compartments and at low to negligible levels in hematopoietic systems [115–117]. This points to the importance of LMNA expression in the maintenance of the differentiated cell state [115]. In HGPS, it has been suggested that stem-cell-driven tissue regeneration may be reduced and tissue-specific differences in apoptosis or regenerative potential may give rise to the tissue-specific segmental aging pattern [118]. Any treatment developed to therapeutically alter the Progerin/LMNA ratio should therefore consider the potential for distinctive role(s) for Lamin A in different tissue compartments.
Conclusions
In conclusion, our analysis describes a range of potential therapeutic approaches that may be used to modulate the expression and accumulation of defective lamin A and/or modify its downstream effects in laminopathies and age-related diseases. However, careful evaluations of these approaches and the potential side effects of drug treatments discussed here are required before consideration as therapeutic treatments in a clinical setting.
References
Dechat T, Adam SA, Taimen P, Shimi T, Goldman RD (2010) Nuclear lamins. Cold Spring Harb Perspect Biol 2:a000547
Dechat T, Gesson K, Foisner R (2010) Lamina-independent lamins in the nuclear interior serve important functions. Cold Spring Harb Symp Quant Biol 75:533–543
Silva L, Cliffe A, Chang L, Knipe DM (2008) Role for A-type lamins in herpesviral DNA targeting and heterochromatin modulation. PLoS Pathog 4:e1000071
Peter A, Stick R (2012) Evolution of the lamin protein family: what introns can tell. Nucleus 3(1):44–59
Lutz RJ, Trujillo MA, Denham KS, Wenger L, Sinensky M (1992) Nucleoplasmic localization of prelamin A: implications for prenylation-dependent lamin A assembly into the nuclear lamina. Proc Natl Acad Sci U S A 89:3000–3004
Pendas AM, Zhou Z, Cadinanos J, Freije JM, Wang J, Hultenby K, Astudillo A, Wernerson A, Rodriguez F, Tryggvason K et al (2002) Defective prelamin A processing and muscular and adipocyte alterations in Zmpste24 metalloproteinase-deficient mice. Nat Genet 31:94–99
Columbaro M, Mattioli E, Schena E, Capanni C, Cenni V, Levy N, Navarro CL, Del Coco R, Squarzoni S, Camozzi D et al (2010) Prelamin A processing and functional effects in restrictive dermopathy. Cell Cycle 9:4766–4768
Reddy S, Comai L (2012) Lamin A, farnesylation and aging. Exp Cell Res 318:1–7
Cao K, Blair CD, Faddah DA, Kieckhaefer JE, Olive M, Erdos MR, Nabel EG, Collins FS (2011) Progerin and telomere dysfunction collaborate to trigger cellular senescence in normal human fibroblasts. J Clin Invest 121:2833–2844
Pollex RL, Hegele RA (2004) Hutchinson–Gilford progeria syndrome. Clin Genet 66:375–381
Eriksson M, Brown WT, Gordon LB, Glynn MW, Singer J, Scott L, Erdos MR, Robbins CM, Moses TY, Berglund P et al (2003) Recurrent de novo point mutations in lamin A cause Hutchinson–Gilford progeria syndrome. Nature 423:293–298
Lopez-Mejia IC, Vautrot V, De Toledo M, Behm-Ansmant I, Bourgeois CF, Navarro CL, Osorio FG, Freije JM, Stevenin J, De Sandre-Giovannoli A et al (2011) A conserved splicing mechanism of the LMNA gene controls premature aging. Hum Mol Genet 20:4540–4555
Moulson CL, Fong LG, Gardner JM, Farber EA, Go G, Passariello A, Grange DK, Young SG, Miner JH (2007) Increased progerin expression associated with unusual LMNA mutations causes severe progeroid syndromes. Hum Mutat 28:882–889
Reunert J, Wentzell R, Walter M, Jakubiczka S, Zenker M, Brune T, Rust S, Marquardt T (2012) Neonatal progeria: increased ratio of progerin to lamin A leads to progeria of the newborn. Eur J Hum Genet 20(9):933–937
Kelley JB, Datta S, Snow CJ, Chatterjee M, Ni L, Spencer A, Yang CS, Cubenas-Potts C, Matunis MJ, Paschal BM (2011) The defective nuclear lamina in Hutchinson-gilford progeria syndrome disrupts the nucleocytoplasmic Ran gradient and inhibits nuclear localization of Ubc9. Mol Cell Biol 31:3378–3395
Goldman RD, Shumaker DK, Erdos MR, Eriksson M, Goldman AE, Gordon LB, Gruenbaum Y, Khuon S, Mendez M, Varga R et al (2004) Accumulation of mutant lamin A causes progressive changes in nuclear architecture in Hutchinson–Gilford progeria syndrome. Proc Natl Acad Sci U S A 101:8963–8968
Burtner CR, Kennedy BK (2010) Progeria syndromes and ageing: what is the connection? Nat Rev Mol Cell Biol 11:567–578
de Magalhaes JP, Budovsky A, Lehmann G, Costa J, Li Y, Fraifeld V, Church GM (2009) The Human Ageing Genomic Resources: online databases and tools for biogerontologists. Aging Cell 8:65–72
de Magalhaes JP, Wuttke D, Wood SH, Plank M, Vora C (2012) Genome–environment interactions that modulate aging: powerful targets for drug discovery. Pharmacol Rev 64:88–101
Scaffidi P, Misteli T (2006) Lamin A-dependent nuclear defects in human aging. Science 312:1059–1063
Ragnauth CD, Warren DT, Liu Y, McNair R, Tajsic T, Figg N, Shroff R, Skepper J, Shanahan CM (2010) Prelamin A acts to accelerate smooth muscle cell senescence and is a novel biomarker of human vascular aging. Circulation 121:2200–2210
Candelario J, Borrego S, Reddy S, Comai L (2011) Accumulation of distinct prelamin A variants in human diploid fibroblasts differentially affects cell homeostasis. Exp Cell Res 317:319–329
Hegele RA, Huff MW, Young TK (2001) Common genomic variation in LMNA modulates indexes of obesity in Inuit. J Clin Endocrinol Metab 86:2747–2751
Weyer C, Wolford JK, Hanson RL, Foley JE, Tataranni PA, Bogardus C, Pratley RE (2001) Subcutaneous abdominal adipocyte size, a predictor of type 2 diabetes, is linked to chromosome 1q21–q23 and is associated with a common polymorphism in LMNA in Pima Indians. Mol Genet Metab 72:231–238
Wolford JK, Hanson RL, Bogardus C, Prochazka M (2001) Analysis of the lamin A/C gene as a candidate for type II diabetes susceptibility in Pima Indians. Diabetologia 44:779–782
Murase Y, Yagi K, Katsuda Y, Asano A, Koizumi J, Mabuchi H (2002) An LMNA variant is associated with dyslipidemia and insulin resistance in the Japanese. Metabolism 51:1017–1021
Steinle NI, Kazlauskaite R, Imumorin IG, Hsueh WC, Pollin TI, O’Connell JR, Mitchell BD, Shuldiner AR (2004) Variation in the lamin A/C gene: associations with metabolic syndrome. Arterioscler Thromb Vasc Biol 24:1708–1713
Liang H, Murase Y, Katuta Y, Asano A, Kobayashi J, Mabuchi H (2005) Association of LMNA 1908C/T polymorphism with cerebral vascular disease and diabetic nephropathy in Japanese men with type 2 diabetes. Clin Endocrinol (Oxf) 63:317–322
Owen KR, Groves CJ, Hanson RL, Knowler WC, Shuldiner AR, Elbein SC, Mitchell BD, Froguel P, Ng MC, Chan JC et al (2007) Common variation in the LMNA gene (encoding lamin A/C) and type 2 diabetes: association analyses in 9,518 subjects. Diabetes 56:879–883
Mesa JL, Loos RJ, Franks PW, Ong KK, Luan J, O’Rahilly S, Wareham NJ, Barroso I (2007) Lamin A/C polymorphisms, type 2 diabetes, and the metabolic syndrome: case–control and quantitative trait studies. Diabetes 56:884–889
Wegner L, Andersen G, Sparso T, Grarup N, Glumer C, Borch-Johnsen K, Jorgensen T, Hansen T, Pedersen O (2007) Common variation in LMNA increases susceptibility to type 2 diabetes and associates with elevated fasting glycemia and estimates of body fat and height in the general population: studies of 7,495 Danish whites. Diabetes 56:694–698
Duesing K, Charpentier G, Marre M, Tichet J, Hercberg S, Froguel P, Gibson F (2008) Evaluating the association of common LMNA variants with type 2 diabetes and quantitative metabolic phenotypes in French Europids. Diabetologia 51:76–81
Akasaka H, Katsuya T, Saitoh S, Sugimoto K, Ohnishi H, Congrains A, Ohnishi M, Ohishi M, Rakugi H, Ogihara T et al (2009) A promoter polymorphism of lamin A/C gene is an independent genetic predisposition to arterial stiffness in a Japanese general population (the Tanno and Sobetsu study). J Atheroscler Thromb 16:404–409
Urbanek M, Nampiaparampil G, D’Souza J, Sefton E, Ackerman C, Legro RS, Dunaif A (2009) The role of genetic variation in the lamin a/c gene in the etiology of polycystic ovary syndrome. J Clin Endocrinol Metab 94:2665–2669
Fontaine-Bisson B, Alessi MC, Saut N, Fumeron F, Marre M, Dutour A, Badens C, Levy N, Tichet J, Juhan-Vague I et al (2010) Polymorphisms of the lamina maturation pathway and their association with the metabolic syndrome: the DESIR prospective study. J Mol Med (Berl) 88:193–201
Halaschek-Wiener J, Amirabbasi-Beik M, Monfared N, Pieczyk M, Sailer C, Kollar A, Thomas R, Agalaridis G, Yamada S, Oliveira L et al (2009) Genetic variation in healthy oldest-old. PLoS One 4:e6641
Yeh HL, Hou SJ, Yen FC, Hong CJ, Liou YJ, Yang AC, Liu ME, Tsai SJ (2011) Polymorphisms in LMNA and near a SERPINA13 gene are not associated with cognitive performance in Chinese elderly males without dementia. Neurosci Lett 504:32–34
Conneely KN, Capell BC, Erdos MR, Sebastiani P, Solovieff N, Swift AJ, Baldwin CT, Budagov T, Barzilai N, Atzmon G et al (2012) Human longevity and common variations in the LMNA gene: a meta-analysis. Aging Cell 11(3):475–481
Lin F, Worman HJ (1993) Structural organization of the human gene encoding nuclear lamin A and nuclear lamin C. J Biol Chem 268:16321–16326
Rodriguez S, Eriksson M (2011) Low and high expressing alleles of the LMNA gene: implications for laminopathy disease development. PLoS One 6:e25472
The International HapMap Consortium (2003) The International HapMap Project. Nature 426:789–796
Chen CY, Chi YH, Mutalif RA, Starost MF, Myers TG, Anderson SA, Stewart CL, Jeang KT (2012) Accumulation of the inner nuclear envelope protein sun1 is pathogenic in progeric and dystrophic laminopathies. Cell 149:565–577
Kenyon CJ (2010) The genetics of ageing. Nature 464:504–512
Ugalde AP, Marino G, Lopez-Otin C (2010) Rejuvenating somatotropic signaling: a therapeutical opportunity for premature aging? Aging (Albany NY) 2:1017–1022
Marino G, Ugalde AP, Fernandez AF, Osorio FG, Fueyo A, Freije JM, Lopez-Otin C (2010) Insulin-like growth factor 1 treatment extends longevity in a mouse model of human premature aging by restoring somatotroph axis function. Proc Natl Acad Sci U S A 107:16268–16273
Arnaldez FI, Helman LJ (2012) Targeting the insulin growth factor receptor 1. Hematol Oncol Clin N Am 26:527–542, vii-viii
Espada J, Varela I, Flores I, Ugalde AP, Cadinanos J, Pendas AM, Stewart CL, Tryggvason K, Blasco MA, Freije JM et al (2008) Nuclear envelope defects cause stem cell dysfunction in premature-aging mice. J Cell Biol 181:27–35
Scaffidi P, Misteli T (2008) Lamin A-dependent misregulation of adult stem cells associated with accelerated ageing. Nat Cell Biol 10:452–459
Meshorer E, Gruenbaum Y (2008) Gone with the Wnt/Notch: stem cells in laminopathies, progeria, and aging. J Cell Biol 181:9–13
Maekawa Y, Minato Y, Ishifune C, Kurihara T, Kitamura A, Kojima H, Yagita H, Sakata-Yanagimoto M, Saito T, Taniuchi I et al (2008) Notch2 integrates signaling by the transcription factors RBP-J and CREB1 to promote T cell cytotoxicity. Nat Immunol 9:1140–1147
Kopan R (ed) (2010) Notch signalling, 1st edn. Waltham, Academic Press
Richards SA, Muter J, Ritchie P, Lattanzi G, Hutchison CJ (2011) The accumulation of un-repairable DNA damage in laminopathy progeria fibroblasts is caused by ROS generation and is prevented by treatment with N-acetyl cysteine. Hum Mol Genet 20:3997–4004
Constantinescu D, Csoka AB, Navara CS, Schatten GP (2010) Defective DSB repair correlates with abnormal nuclear morphology and is improved with FTI treatment in Hutchinson–Gilford progeria syndrome fibroblasts. Exp Cell Res 316:2747–2759
Podmore ID, Griffiths HR, Herbert KE, Mistry N, Mistry P, Lunec J (1998) Vitamin C exhibits pro-oxidant properties. Nature 392:559
Cai L, Koropatnick J, Cherian MG (2001) Roles of vitamin C in radiation-induced DNA damage in presence and absence of copper. Chem Biol Interact 137:75–88
Guidarelli A, De Sanctis R, Cellini B, Fiorani M, Dacha M, Cantoni O (2001) Intracellular ascorbic acid enhances the DNA single-strand breakage and toxicity induced by peroxynitrite in U937 cells. Biochem J 356:509–513
Benson EK, Lee SW, Aaronson SA (2010) Role of progerin-induced telomere dysfunction in HGPS premature cellular senescence. J Cell Sci 123:2605–2612
de Jesus BB, Schneeberger K, Vera E, Tejera A, Harley CB, Blasco MA (2011) The telomerase activator TA-65 elongates short telomeres and increases health span of adult/old mice without increasing cancer incidence. Aging Cell 10:604–621
Mehta IS, Bridger JM, Kill IR (2010) Progeria, the nucleolus and farnesyltransferase inhibitors. Biochem Soc Trans 38:287–291
Marji J, O’Donoghue SI, McClintock D, Satagopam VP, Schneider R, Ratner D, Worman HJ, Gordon LB, Djabali K (2010) Defective lamin A-Rb signaling in Hutchinson–Gilford progeria syndrome and reversal by farnesyltransferase inhibition. PLoS One 5:e11132
Heijink DM, Fehrmann RS, de Vries EG, Koornstra JJ, Oosterhuis D, van der Zee AG, Kleibeuker JH, de Jong S (2011) A bioinformatical and functional approach to identify novel strategies for chemoprevention of colorectal cancer. Oncogene 30:2026–2036
Dechat T, Shimi T, Adam SA, Rusinol AE, Andres DA, Spielmann HP, Sinensky MS, Goldman RD (2007) Alterations in mitosis and cell cycle progression caused by a mutant lamin A known to accelerate human aging. Proc Natl Acad Sci U S A 104:4955–4960
Bridger JM, Kill IR (2004) Aging of Hutchinson–Gilford progeria syndrome fibroblasts is characterised by hyperproliferation and increased apoptosis. Exp Gerontol 39:717–724
Cao K, Graziotto JJ, Blair CD, Mazzulli JR, Erdos MR, Krainc D, Collins FS (2011) Rapamycin reverses cellular phenotypes and enhances mutant protein clearance in Hutchinson–Gilford progeria syndrome cells. Sci Transl Med 3:89ra58
Blagosklonny MV (2011) Progeria, rapamycin and normal aging: recent breakthrough. Aging (Albany NY) 3:685–691
Cenni V, Capanni C, Columbaro M, Ortolani M, D’Apice MR, Novelli G, Fini M, Marmiroli S, Scarano E, Maraldi NM et al (2011) Autophagic degradation of farnesylated prelamin A as a therapeutic approach to lamin-linked progeria. Eur J Histochem 55:e36
Rose C, Menzies FM, Renna M, Acevedo-Arozena A, Corrochano S, Sadiq O, Brown SD, Rubinsztein DC (2010) Rilmenidine attenuates toxicity of polyglutamine expansions in a mouse model of Huntington’s disease. Hum Mol Genet 19:2144–2153
Rajawat Y, Hilioti Z, Bossis I (2010) Autophagy: a target for retinoic acids. Autophagy 6:1224–1226
Rajawat Y, Hilioti Z, Bossis I (2011) Retinoic acid induces autophagosome maturation through redistribution of the cation-independent mannose-6-phosphate receptor. Antioxid Redox Signal 14:2165–2177
Kawakita H, Tsuchida A, Miyazawa K, Naito M, Shigoka M, Kyo B, Enomoto M, Wada T, Katsumata K, Ohyashiki K et al (2009) Growth inhibitory effects of vitamin K2 on colon cancer cell lines via different types of cell death including autophagy and apoptosis. Int J Mol Med 23:709–716
Stravopodis DJ, Karkoulis PK, Konstantakou EG, Melachroinou S, Lampidonis AD, Anastasiou D, Kachrilas S, Messini-Nikolaki N, Papassideri IS, Aravantinos G et al (2009) Grade-dependent effects on cell cycle progression and apoptosis in response to doxorubicin in human bladder cancer cell lines. Int J Oncol 34:137–160
Hundeshagen P, Hamacher-Brady A, Eils R, Brady NR (2011) Concurrent detection of autolysosome formation and lysosomal degradation by flow cytometry in a high-content screen for inducers of autophagy. BMC Biol 9:38
Lee SA, Jung M (2007) The nucleoside analog sangivamycin induces apoptotic cell death in breast carcinoma MCF7/adriamycin-resistant cells via protein kinase Cdelta and JNK activation. J Biol Chem 282:15271–15283
Siman R, Flood DG, Thinakaran G, Neumar RW (2001) Endoplasmic reticulum stress-induced cysteine protease activation in cortical neurons: effect of an Alzheimer’s disease-linked presenilin-1 knock-in mutation. J Biol Chem 276:44736–44743
Arnt CR, Chiorean MV, Heldebrant MP, Gores GJ, Kaufmann SH (2002) Synthetic Smac/DIABLO peptides enhance the effects of chemotherapeutic agents by binding XIAP and cIAP1 in situ. J Biol Chem 277:44236–44243
Gunaratne R, Braucht DW, Rinschen MM, Chou CL, Hoffert JD, Pisitkun T, Knepper MA (2010) Quantitative phosphoproteomic analysis reveals cAMP/vasopressin-dependent signaling pathways in native renal thick ascending limb cells. Proc Natl Acad Sci U S A 107:15653–15658
Georgatos SD, Stournaras C, Blobel G (1988) Heterotypic and homotypic associations between the nuclear lamins: site-specificity and control by phosphorylation. Proc Natl Acad Sci U S A 85:4325–4329
Feng X, Jiang Y, Meltzer P, Yen PM (2000) Thyroid hormone regulation of hepatic genes in vivo detected by complementary DNA microarray. Mol Endocrinol 14:947–955
Kivuva EC, Parker MJ, Cohen MC, Wagner BE, Sobey G (2008) De Barsy syndrome: a review of the phenotype. Clin Dysmorphol 17:99–107
Ioan D, Dumitriu L, Belengeanu V, Bistriceanu M, Maximilian C (1988) Leprechaunism: report of two cases and review. Endocrinologie 26:205–209
Suzuki S, Nishio S, Takeda T, Komatsu M (2012) Gender-specific regulation of response to thyroid hormone in aging. Thyroid Res 5:1
Faggiano A, Del Prete M, Marciello F, Marotta V, Ramundo V, Colao A (2011) Thyroid diseases in elderly. Minerva Endocrinol 36:211–231
Tullai JW, Schaffer ME, Mullenbrock S, Kasif S, Cooper GM (2004) Identification of transcription factor binding sites upstream of human genes regulated by the phosphatidylinositol 3-kinase and MEK/ERK signaling pathways. J Biol Chem 279:20167–20177
Mitsuhashi H, Hayashi YK, Matsuda C, Noguchi S, Wakatsuki S, Araki T, Nishino I (2010) Specific phosphorylation of Ser458 of A-type lamins in LMNA-associated myopathy patients. J Cell Sci 123:3893–3900
Osorio FG, Varela I, Lara E, Puente XS, Espada J, Santoro R, Freije JM, Fraga MF, Lopez-Otin C (2010) Nuclear envelope alterations generate an aging-like epigenetic pattern in mice deficient in Zmpste24 metalloprotease. Aging Cell 9:947–957
Krishnan V, Chow MZ, Wang Z, Zhang L, Liu B, Liu X, Zhou Z (2011) Histone H4 lysine 16 hypoacetylation is associated with defective DNA repair and premature senescence in Zmpste24-deficient mice. Proc Natl Acad Sci U S A 108:12325–12330
Adjei AA, Erlichman C, Davis JN, Cutler DL, Sloan JA, Marks RS, Hanson LJ, Svingen PA, Atherton P, Bishop WR et al (2000) A Phase I trial of the farnesyl transferase inhibitor SCH66336: evidence for biological and clinical activity. Cancer Res 60:1871–1877
Capell BC, Olive M, Erdos MR, Cao K, Faddah DA, Tavarez UL, Conneely KN, Qu X, San H, Ganesh SK et al (2008) A farnesyltransferase inhibitor prevents both the onset and late progression of cardiovascular disease in a progeria mouse model. Proc Natl Acad Sci U S A 105:15902–15907
Varga R, Eriksson M, Erdos MR, Olive M, Harten I, Kolodgie F, Capell BC, Cheng J, Faddah D, Perkins S et al (2006) Progressive vascular smooth muscle cell defects in a mouse model of Hutchinson–Gilford progeria syndrome. Proc Natl Acad Sci U S A 103:3250–3255
Capell BC, Collins FS, Nabel EG (2007) Mechanisms of cardiovascular disease in accelerated aging syndromes. Circ Res 101:13–26
Stehbens WE, Delahunt B, Shozawa T, Gilbert-Barness E (2001) Smooth muscle cell depletion and collagen types in progeric arteries. Cardiovasc Pathol 10:133–136
Yang SH, Meta M, Qiao X, Frost D, Bauch J, Coffinier C, Majumdar S, Bergo MO, Young SG, Fong LG (2006) A farnesyltransferase inhibitor improves disease phenotypes in mice with a Hutchinson–Gilford progeria syndrome mutation. J Clin Invest 116:2115–2121
Yang SH, Qiao X, Farber E, Chang SY, Fong LG, Young SG (2008) Eliminating the synthesis of mature lamin A reduces disease phenotypes in mice carrying a Hutchinson–Gilford progeria syndrome allele. J Biol Chem 283:7094–7099
Fong LG, Frost D, Meta M, Qiao X, Yang SH, Coffinier C, Young SG (2006) A protein farnesyltransferase inhibitor ameliorates disease in a mouse model of progeria. Science 311:1621–1623
Varela I, Pereira S, Ugalde AP, Navarro CL, Suarez MF, Cau P, Cadinanos J, Osorio FG, Foray N, Cobo J et al (2008) Combined treatment with statins and aminobisphosphonates extends longevity in a mouse model of human premature aging. Nat Med 14:767–772
Osorio FG, Obaya AJ, Lopez-Otin C, Freije JM (2009) Accelerated ageing: from mechanism to therapy through animal models. Transgenic Res 18:7–15
Verstraeten VL, Peckham LA, Olive M, Capell BC, Collins FS, Nabel EG, Young SG, Fong LG, Lammerding J (2011) Protein farnesylation inhibitors cause donut-shaped cell nuclei attributable to a centrosome separation defect. Proc Natl Acad Sci U S A 108:4997–5002
Yang SH, Chang SY, Ren S, Wang Y, Andres DA, Spielmann HP, Fong LG, Young SG (2011) Absence of progeria-like disease phenotypes in knock-in mice expressing a non-farnesylated version of progerin. Hum Mol Genet 20:436–444
Fong LG, Vickers TA, Farber EA, Choi C, Yun UJ, Hu Y, Yang SH, Coffinier C, Lee R, Yin L et al (2009) Activating the synthesis of progerin, the mutant prelamin A in Hutchinson–Gilford progeria syndrome, with antisense oligonucleotides. Hum Mol Genet 18:2462–2471
Osorio FG, Navarro CL, Cadinanos J, Lopez-Mejia IC, Quiros PM, Bartoli C, Rivera J, Tazi J, Guzman G, Varela I et al (2011) Splicing-directed therapy in a new mouse model of human accelerated aging. Sci Transl Med 3:106ra107
Jung HJ, Coffinier C, Choe Y, Beigneux AP, Davies BS, Yang SH, Barnes RH 2nd, Hong J, Sun T, Pleasure SJ et al (2012) Regulation of prelamin A but not lamin C by miR-9, a brain-specific microRNA. Proc Natl Acad Sci U S A 109:E423–E431
Weidenfeld I (2012) Inducible microRNA-mediated knockdown of the endogenous human lamin A/C gene. Methods Mol Biol 815:289–305
Sylvius N, Bonne G, Straatman K, Reddy T, Gant TW, Shackleton S (2011) MicroRNA expression profiling in patients with lamin A/C-associated muscular dystrophy. FASEB J 25:3966–3978
Gagne JP, Hunter JM, Labrecque B, Chabot B, Poirier GG (2003) A proteomic approach to the identification of heterogeneous nuclear ribonucleoproteins as a new family of poly(ADP-ribose)-binding proteins. Biochem J 371:331–340
Pumford NR, Halmes NC (1997) Protein targets of xenobiotic reactive intermediates. Annu Rev Pharmacol Toxicol 37:91–117
Butler DC, Messer A (2011) Bifunctional anti-huntingtin proteasome-directed intrabodies mediate efficient degradation of mutant huntingtin exon 1 protein fragments. PLoS One 6:e29199
Kiefer JC (2011) Primer and interviews: advances in targeted gene modification. Dev Dyn 240:2688–2696
Yanez-Munoz RJ, Balaggan KS, MacNeil A, Howe SJ, Schmidt M, Smith AJ, Buch P, MacLaren RE, Anderson PN, Barker SE et al (2006) Effective gene therapy with nonintegrating lentiviral vectors. Nat Med 12:348–353
Tani J, Faustine SJT (2011) Updates on current advances in gene therapy. West Indian Med J 60:188–194
Li JM, Wang YY, Zhao MX, Tan CP, Li YQ, Le XY, Ji LN, Mao ZW (2012) Multifunctional QD-based co-delivery of siRNA and doxorubicin to HeLa cells for reversal of multidrug resistance and real-time tracking. Biomaterials 33:2780–2790
Ashley CE, Carnes EC, Phillips GK, Padilla D, Durfee PN, Brown PA, Hanna TN, Liu J, Phillips B, Carter MB et al (2011) The targeted delivery of multicomponent cargos to cancer cells by nanoporous particle-supported lipid bilayers. Nat Mater 10:389–397
Graziotto JJ, Cao K, Collins FS, Krainc D (2012) Rapamycin activates autophagy in Hutchinson–Gilford progeria syndrome: implications for normal aging and age-dependent neurodegenerative disorders. Autophagy 8:147–151
Children’s Hospital Boston. Study of zoledronic acid, pravastatin, and lonafarnib for patients with progeria. In: ClinicalTrials.gov [Internet]. Bethesda (MD): National Library of Medicine (US). 2009-ongoing [cited 2012 Oct 05]. Available from: http://clinicaltrials.gov/ct2/show/NCT00916747; NLM Identifier: NCT00916747.
Assistance Publique Hopitaux De Marseille: Treatment of the Hutchinson-Gilford Progeria Syndrome With a Combination of Pravastatin and Zoledronic Acid, In: ClinicalTrials.gov [Internet]. Bethesda (MD): National Library of Medicine (US). 2009-ongoing [cited 2012 Oct 05]. Available from: http://clinicaltrials.gov/ct2/show/NCT00731016; NLM Identifier: NCT00731016.
Constantinescu D, Gray HL, Sammak PJ, Schatten GP, Csoka AB (2006) Lamin A/C expression is a marker of mouse and human embryonic stem cell differentiation. Stem Cells 24:177–185
Rober RA, Sauter H, Weber K, Osborn M (1990) Cells of the cellular immune and hemopoietic system of the mouse lack lamins A/C: distinction versus other somatic cells. J Cell Sci 95(Pt 4):587–598
Rober RA, Weber K, Osborn M (1989) Differential timing of nuclear lamin A/C expression in the various organs of the mouse embryo and the young animal: a developmental study. Development 105:365–378
Halaschek-Wiener J, Brooks-Wilson A (2007) Progeria of stem cells: stem cell exhaustion in Hutchinson–Gilford progeria syndrome. J Gerontol A Biol Sci Med Sci 62:3–8
Wu G, Lyapina S, Das I, Li J, Gurney M, Pauley A, Chui I, Deshaies RJ, Kitajewski J (2001) SEL-10 is an inhibitor of notch signaling that targets notch for ubiquitin-mediated protein degradation. Mol Cell Biol 21:7403–7415
Zhang CP, Yang JL, Zhang J, Li L, Huang L, Ji SY, Hu ZY, Gao F, Liu YX (2011) Notch signaling is involved in ovarian follicle development by regulating granulosa cell proliferation. Endocrinology 152:2437–2447
Metacore from GeneGo Inc., A Thompson Reuters company, 2012 (http://www.genego.com)
Shih IM, Wang TL (2007) Notch signaling, gamma-secretase inhibitors, and cancer therapy. Cancer Res 67:1879–1882
Chernoff AE, Granowitz EV, Shapiro L, Vannier E, Lonnemann G, Angel JB, Kennedy JS, Rabson AR, Wolff SM, Dinarello CA (1995) A randomized, controlled trial of IL-10 in humans. Inhibition of inflammatory cytokine production and immune responses. J Immunol 154:5492–5499
Tomita T, Iwatsubo T (2004) The inhibition of gamma-secretase as a therapeutic approach to Alzheimer’s disease. Drug News Perspect 17:321–325
Bekaii-Saab TS, Mortazavi A, Hicks LG, Zalupski M, Pelley RJ, Chan KK, Kraut EH (2006) A phase II study of chloroquinoxaline sulfonamide (CQS) in patients with metastatic colorectal carcinoma (MCRC). Investig New Drugs 24:343–346
Wanka J, Jones LI, Wood PH, Dixon AS (1964) Indomethacin in rheumatic diseases. a controlled clinical trial. Ann Rheum Dis 23:218–225
Takebe N, Warren RQ, Ivy SP (2011) Breast cancer growth and metastasis: interplay between cancer stem cells, embryonic signaling pathways and epithelial-to-mesenchymal transition. Breast Cancer Res 13:211
Scherr DS, Pitts WR Jr (2003) The nonsteroidal effects of diethylstilbestrol: the rationale for androgen deprivation therapy without estrogen deprivation in the treatment of prostate cancer. J Urol 170:1703–1708
Huynh C, Poliseno L, Segura MF, Medicherla R, Haimovic A, Menendez S, Shang S, Pavlick A, Shao Y, Darvishian F et al (2011) The novel gamma secretase inhibitor RO4929097 reduces the tumor initiating potential of melanoma. PLoS One 6:e25264
Yamada J, Yoshimura S, Yamakawa H, Sawada M, Nakagawa M, Hara S, Kaku Y, Iwama T, Naganawa T, Banno Y et al (2003) Cell permeable ROS scavengers, Tiron and Tempol, rescue PC12 cell death caused by pyrogallol or hypoxia/reoxygenation. Neurosci Res 45:1–8
Han YH, Park WH (2009) Tiron, a ROS scavenger, protects human lung cancer Calu-6 cells against antimycin A-induced cell death. Oncol Rep 21:253–261
Stanley JL, Andersson IJ, Hirt CJ, Moore L, Dilworth MR, Chade AR, Sibley CP, Davidge ST, Baker PN (2012) Effect of the anti-oxidant tempol on fetal growth in a mouse model of fetal growth restriction. Biol Reprod
Lee S, Pagoria D, Raigrodski A, Geurtsen W (2007) Effects of combinations of ROS scavengers on oxidative DNA damage caused by visible-light-activated camphorquinone/N,N-dimethyl-p-toluidine. J Biomed Mater Res B Appl Biomater 83:391–399
Stey C, Steurer J, Bachmann S, Medici TC, Tramer MR (2000) The effect of oral N-acetylcysteine in chronic bronchitis: a quantitative systematic review. Eur Respir J 16:253–262
Anonymous (1974) Toxicological evaluation of some food additives including anticaking agents, antimicrobials, antioxidants, emulsifiers and thickening agents. FAO Nutr Meet Rep Ser: 1–520
Jialal I, Grundy SM (1992) Effect of dietary supplementation with alpha-tocopherol on the oxidative modification of low density lipoprotein. J Lipid Res 33:899–906
Chan AS, Cheung MC, Law SC, Chan JH (2004) Phase II study of alpha-tocopherol in improving the cognitive function of patients with temporal lobe radionecrosis. Cancer 100:398–404
Fuks B, Talaga P, Huart C, Henichart JP, Bertrand K, Grimee R, Lorent G (2005) In vitro properties of 5-(benzylsulfonyl)-4-bromo-2-methyl-3(2H)-pyridazinone: a novel permeability transition pore inhibitor. Eur J Pharmacol 519:24–30
Grohm J, Plesnila N, Culmsee C (2010) Bid mediates fission, membrane permeabilization and peri-nuclear accumulation of mitochondria as a prerequisite for oxidative neuronal cell death. Brain Behav Immun 24:831–838
Becattini B, Sareth S, Zhai D, Crowell KJ, Leone M, Reed JC, Pellecchia M (2004) Targeting apoptosis via chemical design: inhibition of bid-induced cell death by small organic molecules. Chem Biol 11:1107–1117
Wei MC, Lindsten T, Mootha VK, Weiler S, Gross A, Ashiya M, Thompson CB, Korsmeyer SJ (2000) tBID, a membrane-targeted death ligand, oligomerizes BAK to release cytochrome c. Genes Dev 14:2060–2071
Kimura H, Sato Y, Tajima Y, Suzuki H, Yukitake H, Imaeda T, Kajino M, Oki H, Takizawa M, Tanida S (2010) BTZO-1, a cardioprotective agent, reveals that macrophage migration inhibitory factor regulates ARE-mediated gene expression. Chem Biol 17:1282–1294
Furlong IJ, Lopez Mediavilla C, Ascaso R, Lopez Rivas A, Collins MK (1998) Induction of apoptosis by valinomycin: mitochondrial permeability transition causes intracellular acidification. Cell Death Differ 5:214–221
Mastrangelo AJ, Zou S, Hardwick JM, Betenbaugh MJ (1999) Antiapoptosis chemicals prolong productive lifetimes of mammalian cells upon Sindbis virus vector infection. Biotechnol Bioeng 65:298–305
Dumont A, Hehner SP, Hofmann TG, Ueffing M, Droge W, Schmitz ML (1999) Hydrogen peroxide-induced apoptosis is CD95-independent, requires the release of mitochondria-derived reactive oxygen species and the activation of NF-kappa β. Oncogene 18:747–757
Gross A, McDonnell JM, Korsmeyer SJ (1999) BCL-2 family members and the mitochondria in apoptosis. Genes Dev 13:1899–1911
Scarlett JL, Sheard PW, Hughes G, Ledgerwood EC, Ku HH, Murphy MP (2000) Changes in mitochondrial membrane potential during staurosporine-induced apoptosis in Jurkat cells. FEBS Lett 475:267–272
Pastorino JG, Hoek JB (2000) Ethanol potentiates tumor necrosis factor-alpha cytotoxicity in hepatoma cells and primary rat hepatocytes by promoting induction of the mitochondrial permeability transition. Hepatology 31:1141–1152
Damico R, Simms T, Kim BS, Tekeste Z, Amankwan H, Damarla M, Hassoun PM (2011) p53 mediates cigarette smoke-induced apoptosis of pulmonary endothelial cells: inhibitory effects of macrophage migration inhibitor factor. Am J Respir Cell Mol Biol 44:323–332
Zhao CQ, Zhang YH, Jiang SD, Jiang LS, Dai LY (2010) Both endoplasmic reticulum and mitochondria are involved in disc cell apoptosis and intervertebral disc degeneration in rats. Age (Dordr) 32:161–177
Chu W, Zhang J, Zeng C, Rothfuss J, Tu Z, Chu Y, Reichert DE, Welch MJ, Mach RH (2005) N-benzylisatin sulfonamide analogues as potent caspase-3 inhibitors: synthesis, in vitro activity, and molecular modeling studies. J Med Chem 48:7637–7647
Lee D, Long SA, Murray JH, Adams JL, Nuttall ME, Nadeau DP, Kikly K, Winkler JD, Sung CM, Ryan MD et al (2001) Potent and selective nonpeptide inhibitors of caspases 3 and 7. J Med Chem 44:2015–2026
Hoffstrom BG, Kaplan A, Letso R, Schmid RS, Turmel GJ, Lo DC, Stockwell BR (2010) Inhibitors of protein disulfide isomerase suppress apoptosis induced by misfolded proteins. Nat Chem Biol 6:900–906
Cherqui S (2012) Cysteamine therapy: a treatment for cystinosis, not a cure. Kidney Int 81:127–129
Tsilou ET, Thompson D, Lindblad AS, Reed GF, Rubin B, Gahl W, Thoene J, Del Monte M, Schneider JA, Granet DB et al (2003) A multicentre randomised double masked clinical trial of a new formulation of topical cysteamine for the treatment of corneal cystine crystals in cystinosis. Br J Ophthalmol 87:28–31
Komarova EA, Gudkov AV (2000) Suppression of p53: a new approach to overcome side effects of antitumor therapy. Biochemistry (Mosc) 65:41–48
Komarov PG, Komarova EA, Kondratov RV, Christov-Tselkov K, Coon JS, Chernov MV, Gudkov AV (1999) A chemical inhibitor of p53 that protects mice from the side effects of cancer therapy. Science 285:1733–1737
Strom E, Sathe S, Komarov PG, Chernova OB, Pavlovska I, Shyshynova I, Bosykh DA, Burdelya LG, Macklis RM, Skaliter R et al (2006) Small-molecule inhibitor of p53 binding to mitochondria protects mice from gamma radiation. Nat Chem Biol 2:474–479
Elimadi A, Jullien V, Tillement JP, Morin D (2003) S-15176 inhibits mitochondrial permeability transition via a mechanism independent of its antioxidant properties. Eur J Pharmacol 468:93–101
McCall M, Toso C, Emamaullee J, Pawlick R, Edgar R, Davis J, Maciver A, Kin T, Arch R, Shapiro AM (2011) The caspase inhibitor IDN-6556 (PF3491390) improves marginal mass engraftment after islet transplantation in mice. Surgery 150:48–55
Rao VA, Klein SR, Bonar SJ, Zielonka J, Mizuno N, Dickey JS, Keller PW, Joseph J, Kalyanaraman B, Shacter E (2010) The antioxidant transcription factor Nrf2 negatively regulates autophagy and growth arrest induced by the anticancer redox agent mitoquinone. J Biol Chem 285:34447–34459
Deretic V (2012) Autophagy as an innate immunity paradigm: expanding the scope and repertoire of pattern recognition receptors. Curr Opin Immunol 24:21–31
Vieth R, Chan PC, MacFarlane GD (2001) Efficacy and safety of vitamin D3 intake exceeding the lowest observed adverse effect level. Am J Clin Nutr 73:288–294
Wang Q, Liang B, Shirwany NA, Zou MH (2011) 2-Deoxy-D-glucose treatment of endothelial cells induces autophagy by reactive oxygen species-mediated activation of the AMP-activated protein kinase. PLoS One 6:e17234
Xi H, Kurtoglu M, Liu H, Wangpaichitr M, You M, Liu X, Savaraj N, Lampidis TJ (2011) 2-Deoxy-d-glucose activates autophagy via endoplasmic reticulum stress rather than ATP depletion. Cancer Chemother Pharmacol 67:899–910
Bijnsdorp IV, Peters GJ, Temmink OH, Fukushima M, Kruyt FA (2010) Differential activation of cell death and autophagy results in an increased cytotoxic potential for trifluorothymidine compared to 5-fluorouracil in colon cancer cells. Int J Cancer 126:2457–2468
Lin Q, Gao XS, Qiao XY, Chen K, Wang YD, Zhou ZG (2008) Phase II clinical trial of concurrent chemoradiotherapy (cisplatin plus 5-fluorouracil) for esophageal cancer. Ai Zheng 27:1077–1081
Xia LP, Li LY, Fei XF, Liang ZQ (2010) Autophagy is involved in 6-OHDA-induced dopaminergic cell death. Nan Fang Yi Ke Da Xue Xue Bao 30:2649–2651
Ding WX, Ni HM, Gao W, Hou YF, Melan MA, Chen X, Stolz DB, Shao ZM, Yin XM (2007) Differential effects of endoplasmic reticulum stress-induced autophagy on cell survival. J Biol Chem 282:4702–4710
Balgi AD, Fonseca BD, Donohue E, Tsang TC, Lajoie P, Proud CG, Nabi IR, Roberge M (2009) Screen for chemical modulators of autophagy reveals novel therapeutic inhibitors of mTORC1 signaling. PLoS One 4:e7124
Naccarelli GV, Rinkenberger RL, Dougherty AH, Fitzgerald DM (1989) Adverse effects of amiodarone. Pathogenesis, incidence and management. Med Toxicol Adverse Drug Exp 4:246–253
Cheong H, Lindsten T, Thompson CB (2012) Autophagy and ammonia. Autophagy 8(1):122–123
Goussetis DJ, Altman JK, Glaser H, McNeer JL, Tallman MS, Platanias LC (2010) Autophagy is a critical mechanism for the induction of the antileukemic effects of arsenic trioxide. J Biol Chem 285:29989–29997
Liu SY, Wen CY, Lee YJ, Lee TC (2010) XPC silencing sensitizes glioma cells to arsenic trioxide via increased oxidative damage. Toxicol Sci 116:183–193
Berenson JR, Matous J, Swift RA, Mapes R, Morrison B, Yeh HS (2007) A phase I/II study of arsenic trioxide/bortezomib/ascorbic acid combination therapy for the treatment of relapsed or refractory multiple myeloma. Clin Cancer Res 13:1762–1768
Din FV, Valanciute A, Houde V, Zibrova D, Green KA, Sakamoto K, Alessi DR, Dunlop MG (2012) Aspirin inhibits mTOR signaling, activates AMP-activated protein kinase, and induces autophagy in colorectal cancer cells. Gastroenterology 142(7):1504–15.e3
Sandler RS, Halabi S, Baron JA, Budinger S, Paskett E, Keresztes R, Petrelli N, Pipas JM, Karp DD, Loprinzi CL et al (2003) A randomized trial of aspirin to prevent colorectal adenomas in patients with previous colorectal cancer. N Engl J Med 348:883–890
Zhang Q, Yang YJ, Wang H, Dong QT, Wang TJ, Qian HY, Xu H (2012) Autophagy activationA novel mechanism of atorvastatin to protect mesenchymal stem cells from hypoxia and serum deprivation via AMP-activated protein kinase/mammalian target of rapamycin pathway. Stem Cells Dev 21(8):1321–1332
Newman C, Tsai J, Szarek M, Luo D, Gibson E (2006) Comparative safety of atorvastatin 80 mg versus 10 mg derived from analysis of 49 completed trials in 14,236 patients. Am J Cardiol 97:61–67
Pattingre S, Bauvy C, Codogno P (2003) Amino acids interfere with the ERK1/2-dependent control of macroautophagy by controlling the activation of Raf-1 in human colon cancer HT-29 cells. J Biol Chem 278:16667–16674
Schenone S, Brullo C, Musumeci F, Radi M, Botta M (2011) ATP-competitive inhibitors of mTOR: an update. Curr Med Chem 18:2995–3014
Ariyoshi-Kishino K, Hashimoto K, Amano O, Saitoh J, Kochi M, Sakagami H (2010) Tumor-specific cytotoxicity and type of cell death induced by benzaldehyde. Anticancer Res 30:5069–5076
Jia L, Gopinathan G, Sukumar JT, Gribben JG (2012) Blocking autophagy prevents bortezomib-induced NF-κβ activation by reducing I-κβα degradation in lymphoma cells. PLoS One 7:e32584
Komatsu S, Miyazawa K, Moriya S, Takase A, Naito M, Inazu M, Kohno N, Itoh M, Tomoda A (2012) Clarithromycin enhances bortezomib-induced cytotoxicity via endoplasmic reticulum stress-mediated CHOP (GADD153) induction and autophagy in breast cancer cells. Int J Oncol 40:1029–1039
Phuphanich S, Supko JG, Carson KA, Grossman SA, Burt Nabors L, Mikkelsen T, Lesser G, Rosenfeld S, Desideri S, Olson JJ (2010) Phase 1 clinical trial of bortezomib in adults with recurrent malignant glioma. J Neurooncol 100:95–103
Kim DS, Kim JH, Lee GH, Kim HT, Lim JM, Chae SW, Chae HJ, Kim HR (2010) p38 Mitogen-activated protein kinase is involved in endoplasmic reticulum stress-induced cell death and autophagy in human gingival fibroblasts. Biol Pharm Bull 33:545–549
Bae N, Ahn T, Chung S, Oh MS, Ko H, Oh H, Park G, Yang HO (2011) The neuroprotective effect of modified Yeoldahanso-tang via autophagy enhancement in models of Parkinson’s disease. J Ethnopharmacol 134:313–322
Pfisterer SG, Mauthe M, Codogno P, Proikas-Cezanne T (2011) Ca2+/calmodulin-dependent kinase (CaMK) signaling via CaMKI and AMP-activated protein kinase contributes to the regulation of WIPI-1 at the onset of autophagy. Mol Pharmacol 80:1066–1075
Yoon JH, Ahn SG, Lee BH, Jung SH, Oh SH (2012) Role of autophagy in chemoresistance: regulation of the ATM-mediated DNA-damage signaling pathway through activation of DNA-PKcs and PARP-1. Biochem Pharmacol 83:747–757
Goldfrank LR, Hoffman RS (2007) Goldfrank’s manual of toxicologic emergencies. McGraw-Hill Medical, New York
Kim H, Bernard ME, Flickinger J, Epperly MW, Wang H, Dixon TM, Shields D, Houghton F, Zhang X, Greenberger JS (2011) The autophagy-inducing drug carbamazepine is a radiation protector and mitigator. Int J Radiat Biol 87:1052–1060
Xiong N, Jia M, Chen C, Xiong J, Zhang Z, Huang J, Hou L, Yang H, Cao X, Liang Z et al (2011) Potential autophagy enhancers attenuate rotenone-induced toxicity in SH-SY5Y. Neuroscience 199:292–302
Dam M, Christiansen J, Kristensen CB, Helles A, Jaegerskou A, Schmiegelow M (1981) Carbamazepine: a clinical biopharmaceutical study. Eur J Clin Pharmacol 20:59–64
Pattingre S, Bauvy C, Levade T, Levine B, Codogno P (2009) Ceramide-induced autophagy: to junk or to protect cells? Autophagy 5:558–560
Daido S, Kanzawa T, Yamamoto A, Takeuchi H, Kondo Y, Kondo S (2004) Pivotal role of the cell death factor BNIP3 in ceramide-induced autophagic cell death in malignant glioma cells. Cancer Res 64:4286–4293
Li X, Fan Z (2010) The epidermal growth factor receptor antibody cetuximab induces autophagy in cancer cells by downregulating HIF-1alpha and Bcl-2 and activating the beclin 1/hVps34 complex. Cancer Res 70:5942–5952
Maubec E, Petrow P, Scheer-Senyarich I, Duvillard P, Lacroix L, Gelly J, Certain A, Duval X, Crickx B, Buffard V et al (2011) Phase II study of cetuximab as first-line single-drug therapy in patients with unresectable squamous cell carcinoma of the skin. J Clin Oncol 29:3419–3426
Maycotte P, Aryal S, Cummings CT, Thorburn J, Morgan MJ, Thorburn A (2012) Chloroquine sensitizes breast cancer cells to chemotherapy independent of autophagy. Autophagy 8:200–212
Duenas-Gonzalez A, Zarba JJ, Patel F, Alcedo JC, Beslija S, Casanova L, Pattaranutaporn P, Hameed S, Blair JM, Barraclough H et al (2011) Phase III, open-label, randomized study comparing concurrent gemcitabine plus cisplatin and radiation followed by adjuvant gemcitabine and cisplatin versus concurrent cisplatin and radiation in patients with stage IIB to IVA carcinoma of the cervix. J Clin Oncol 29:1678–1685
Tsvetkov AS, Miller J, Arrasate M, Wong JS, Pleiss MA, Finkbeiner S (2010) A small-molecule scaffold induces autophagy in primary neurons and protects against toxicity in a Huntington disease model. Proc Natl Acad Sci U S A 107:16982–16987
Nsimba SE (2009) Effects of daily chlorpromazine administration on behavioural and physiological parameters in the rat. Indian J Physiol Pharmacol 53:209–218
Campbell GR, Spector SA (2011) Hormonally active vitamin D3 (1alpha,25-dihydroxycholecalciferol) triggers autophagy in human macrophages that inhibits HIV-1 infection. J Biol Chem 286:18890–18902
Vasquez A, Manso G, Cannell J (2004) The clinical importance of vitamin D (cholecalciferol): a paradigm shift with implications for all healthcare providers. Altern Ther Health Med 10:28–36, quiz 37, 94
Williams A, Sarkar S, Cuddon P, Ttofi EK, Saiki S, Siddiqi FH, Jahreiss L, Fleming A, Pask D, Goldsmith P et al (2008) Novel targets for Huntington’s disease in an mTOR-independent autophagy pathway. Nat Chem Biol 4:295–305
Perera PM, Jayamanna SF, Hettiarachchi R, Abeysinghe C, Karunatilake H, Dawson AH, Buckley NA (2009) A phase II clinical trial to assess the safety of clonidine in acute organophosphorus pesticide poisoning. Trials 10:73
Zhang T, Li Y, Park KA, Byun HS, Won M, Jeon J, Lee Y, Seok JH, Choi SW, Lee SH et al (2012) Cucurbitacin induces autophagy through mitochondrial ROS production which counteracts to limit caspase-dependent apoptosis. Autophagy 8(4):559–576
Ferree A, Guillily M, Li H, Smith K, Takashima A, Squillace R, Weigele M, Collins JJ, Wolozin B (2011) Regulation of physiologic actions of LRRK2: focus on autophagy. Neurodegener Dis 10(1–4):238–241
Hartford CM, Desai AA, Janisch L, Karrison T, Rivera VM, Berk L, Loewy JW, Kindler H, Stadler WM, Knowles HL et al (2009) A phase I trial to determine the safety, tolerability, and maximum tolerated dose of deforolimus in patients with advanced malignancies. Clin Cancer Res 15:1428–1434
Vara D, Salazar M, Olea-Herrero N, Guzman M, Velasco G, Diaz-Laviada I (2011) Anti-tumoral action of cannabinoids on hepatocellular carcinoma: role of AMPK-dependent activation of autophagy. Cell Death Differ 18:1099–1111
Molitoris JK, McColl KS, Swerdlow S, Matsuyama M, Lam M, Finkel TH, Matsuyama S, Distelhorst CW (2011) Glucocorticoid elevation of dexamethasone-induced gene 2 (Dig2/RTP801/REDD1) protein mediates autophagy in lymphocytes. J Biol Chem 286:30181–30189
Bjornson CL, Klassen TP, Williamson J, Brant R, Mitton C, Plint A, Bulloch B, Evered L, Johnson DW (2004) A randomized trial of a single dose of oral dexamethasone for mild croup. N Engl J Med 351:1306–1313
Wasko BM, Dudakovic A, Hohl RJ (2011) Bisphosphonates induce autophagy by depleting geranylgeranyl diphosphate. J Pharmacol Exp Ther 337:540–546
Diel IJ, Bergner R, Grotz KA (2007) Adverse effects of bisphosphonates: current issues. J Support Oncol 5:475–482
Jing K, Song KS, Shin S, Kim N, Jeong S, Oh HR, Park JH, Seo KS, Heo JY, Han J et al (2011) Docosahexaenoic acid induces autophagy through p53/AMPK/mTOR signaling and promotes apoptosis in human cancer cells harboring wild-type p53. Autophagy 7:1348–1358
Bedikian AY, DeConti RC, Conry R, Agarwala S, Papadopoulos N, Kim KB, Ernstoff M (2011) Phase 3 study of docosahexaenoic acid-paclitaxel versus dacarbazine in patients with metastatic malignant melanoma. Ann Oncol 22:787–793
Kobayashi S, Volden P, Timm D, Mao K, Xu X, Liang Q (2010) Transcription factor GATA4 inhibits doxorubicin-induced autophagy and cardiomyocyte death. J Biol Chem 285:793–804
Kattan J, Droz JP, Couvreur P, Marino JP, Boutan-Laroze A, Rougier P, Brault P, Vranckx H, Grognet JM, Morge X et al (1992) Phase I clinical trial and pharmacokinetic evaluation of doxorubicin carried by polyisohexylcyanoacrylate nanoparticles. Investig New Drugs 10:191–199
Shen S, Kepp O, Martins I, Vitale I, Souquere S, Castedo M, Pierron G, Kroemer G (2010) Defective autophagy associated with LC3 puncta in epothilone-resistant cancer cells. Cell Cycle 9:377–383
Marino ML, Fais S, Djavaheri-Mergny M, Villa A, Meschini S, Lozupone F, Venturi G, Della Mina P, Pattingre S, Rivoltini L et al (2010) Proton pump inhibition induces autophagy as a survival mechanism following oxidative stress in human melanoma cells. Cell Death Dis 1:e87
Travelli C, Drago V, Maldi E, Kaludercic N, Galli U, Boldorini R, Di Lisa F, Tron GC, Canonico PL, Genazzani AA (2011) Reciprocal potentiation of the antitumoral activities of FK866, an inhibitor of nicotinamide phosphoribosyltransferase, and etoposide or cisplatin in neuroblastoma cells. J Pharmacol Exp Ther 338:829–840
Sessa C, Zucchetti M, Cerny T, Pagani O, Cavalli F, De Fusco M, De Jong J, Gentili D, McDaniel C, Prins C et al (1995) Phase I clinical and pharmacokinetic study of oral etoposide phosphate. J Clin Oncol 13:200–209
Roscic A, Baldo B, Crochemore C, Marcellin D, Paganetti P (2011) Induction of autophagy with catalytic mTOR inhibitors reduces huntingtin aggregates in a neuronal cell model. J Neurochem 119:398–407
Curran MP (2012) Everolimus: in patients with subependymal giant cell astrocytoma associated with tuberous sclerosis complex. Paediatr Drugs 14:51–60
Xia HG, Zhang L, Chen G, Zhang T, Liu J, Jin M, Ma X, Ma D, Yuan J (2010) Control of basal autophagy by calpain1 mediated cleavage of ATG5. Autophagy 6:61–66
Wurthmann C, Klieser E, Lehmann E (1995) Interaction of therapeutic effects and side effects in drug therapy of generalized anxiety disorders with low dosage fluspirilene. Fortschr Neurol Psychiatr 63:72–77
Chouinard G, Annable L, Steinberg S (1986) A controlled clinical trial of fluspirilene, a long-acting injectable neuroleptic, in schizophrenic patients with acute exacerbation. J Clin Psychopharmacol 6:21–26
Ravikumar B, Stewart A, Kita H, Kato K, Duden R, Rubinsztein DC (2003) Raised intracellular glucose concentrations reduce aggregation and cell death caused by mutant huntingtin exon 1 by decreasing mTOR phosphorylation and inducing autophagy. Hum Mol Genet 12:985–994
Lian J, Karnak D, Xu L (2010) The Bcl-2-Beclin 1 interaction in (−)-gossypol-induced autophagy versus apoptosis in prostate cancer cells. Autophagy 6:1201–1203
Gough DR, Cotter TG (2011) Hydrogen peroxide: a Jekyll and Hyde signalling molecule. Cell Death Dis 2:e213
Calabretta B, Salomoni P (2012) Suppression of autophagy by BCR/ABL. Front Biosci (Schol Ed) 4:453–460
Deininger M, Buchdunger E, Druker BJ (2005) The development of imatinib as a therapeutic agent for chronic myeloid leukemia. Blood 105:2640–2653
Christian F, Anthony DF, Vadrevu S, Riddell T, Day JP, McLeod R, Adams DR, Baillie GS, Houslay MD (2010) p62 (SQSTM1) and cyclic AMP phosphodiesterase-4A4 (PDE4A4) locate to a novel, reversible protein aggregate with links to autophagy and proteasome degradation pathways. Cell Signal 22:1576–1596
Lim L, Jackson-Lewis V, Wong LC, Shui GH, Goh AX, Kesavapany S, Jenner AM, Fivaz M, Przedborski S, Wenk MR (2012) Lanosterol induces mitochondrial uncoupling and protects dopaminergic neurons from cell death in a model for Parkinson’s disease. Cell Death Differ 19:416–427
Savaraj N, You M, Wu C, Wangpaichitr M, Kuo MT, Feun LG (2010) Arginine deprivation, autophagy, apoptosis (AAA) for the treatment of melanoma. Curr Mol Med 10:405–412
Haspel J, Shaik RS, Ifedigbo E, Nakahira K, Dolinay T, Englert JA, Choi AM (2011) Characterization of macroautophagic flux in vivo using a leupeptin-based assay. Autophagy 7:629–642
Meng N, Zhao J, Su L, Zhao B, Zhang Y, Zhang S, Miao J (2012) A butyrolactone derivative suppressed lipopolysaccharide-induced autophagic injury through inhibiting the autoregulatory loop of p8 and p53 in vascular endothelial cells. Int J Biochem Cell Biol 44:311–319
Shimada K, Motoi Y, Ishiguro K, Kambe T, Matsumoto SE, Itaya M, Kunichika M, Mori H, Shinohara A, Chiba M et al (2012) Long-term oral lithium treatment attenuates motor disturbance in tauopathy model mice: Implications of autophagy promotion. Neurobiol Dis 46:101–108
Mainguet P, Fiasse R (1977) Double-blind placebo-controlled study of loperamide (Imodium) in chronic diarrhoea caused by ileocolic disease or resection. Gut 18:575–579
Pan Y, Gao Y, Chen L, Gao G, Dong H, Yang Y, Dong B, Chen X (2011) Targeting autophagy augments in vitro and in vivo antimyeloma activity of DNA-damaging chemotherapy. Clin Cancer Res 17:3248–3258
Palumbo A, Bringhen S, Petrucci MT, Musto P, Rossini F, Nunzi M, Lauta VM, Bergonzi C, Barbui A, Caravita T et al (2004) Intermediate-dose melphalan improves survival of myeloma patients aged 50 to 70: results of a randomized controlled trial. Blood 104:3052–3057
Shi WY, Xiao D, Wang L, Dong LH, Yan ZX, Shen ZX, Chen SJ, Chen Y, Zhao WL (2012) Therapeutic metformin/AMPK activation blocked lymphoma cell growth via inhibition of mTOR pathway and induction of autophagy. Cell Death Dis 3:e275
Scarpello JH, Howlett HC (2008) Metformin therapy and clinical uses. Diab Vasc Dis Res 5:157–167
Olsen EA, Dunlap FE, Funicella T, Koperski JA, Swinehart JM, Tschen EH, Trancik RJ (2002) A randomized clinical trial of 5 % topical minoxidil versus 2 % topical minoxidil and placebo in the treatment of androgenetic alopecia in men. J Am Acad Dermatol 47:377–385
Gills JJ, Lopiccolo J, Dennis PA (2008) Nelfinavir, a new anti-cancer drug with pleiotropic effects and many paths to autophagy. Autophagy 4:107–109
Brunner TB, Geiger M, Grabenbauer GG, Lang-Welzenbach M, Mantoni TS, Cavallaro A, Sauer R, Hohenberger W, McKenna WG (2008) Phase I trial of the human immunodeficiency virus protease inhibitor nelfinavir and chemoradiation for locally advanced pancreatic cancer. J Clin Oncol 26:2699–2706
Ofori-Adjei D, Dodoo ANO, Appiah-Danquah A, Couper M (1990) A review of the safety of niclosamide, pyrantel, triclabendazole and oxamniquine. Risk Saf Med 20:113–122
Ghanizadeh A, Haghighat R (2012) Nortriptyline for treating enuresis in ADHD-a randomized double-blind controlled clinical trial. Pediatr Nephrol. doi:10.1007/s00467-012-2211-z
Hu C, Zou MJ, Zhao L, Lu N, Sun YJ, Gou SH, Xi T, Guo QL (2012) E Platinum, a newly synthesized platinum compound, induces autophagy via inhibiting phosphorylation of mTOR in gastric carcinoma BGC-823 cells. Toxicol Lett 210:78–86
Raymond E, Chaney SG, Taamma A, Cvitkovic E (1998) Oxaliplatin: a review of preclinical and clinical studies. Ann Oncol 9:1053–1071
Tong Y, Liu YY, You LS, Qian WB (2012) Perifosine induces protective autophagy and upregulation of ATG5 in human chronic myelogenous leukemia cells in vitro. Acta Pharmacol Sin 33(4):542–552
Vink SR, Schellens JH, Beijnen JH, Sindermann H, Engel J, Dubbelman R, Moppi G, Hillebrand MJ, Bartelink H, Verheij M (2006) Phase I and pharmacokinetic study of combined treatment with perifosine and radiation in patients with advanced solid tumours. Radiother Oncol 80:207–213
Cole PL, Beamer AD, McGowan N, Cantillon CO, Benfell K, Kelly RA, Hartley LH, Smith TW, Antman EM (1990) Efficacy and safety of perhexiline maleate in refractory angina. A double-blind placebo-controlled clinical trial of a novel antianginal agent. Circulation 81:1260–1270
Remijsen Q, Vanden Berghe T, Wirawan E, Asselbergh B, Parthoens E, De Rycke R, Noppen S, Delforge M, Willems J, Vandenabeele P (2011) Neutrophil extracellular trap cell death requires both autophagy and superoxide generation. Cell Res 21:290–304
Chen D, Fan W, Lu Y, Ding X, Chen S, Zhong Q (2012) A mammalian autophagosome maturation mechanism mediated by TECPR1 and the Atg12-Atg5 conjugate. Mol Cell 45:629–641
van Vloten WA (2003) Pimozide: use in dermatology. Dermatol Online J 9:3
Chassan JB (1959) A statistical description of a clinical trial of promazine. Psychiatr Q 33:700–714
Page CB, Duffull SB, Whyte IM, Isbister GK (2009) Promethazine overdose: clinical effects, predicting delirium and the effect of charcoal. QJM 102:123–131
Bahro M, Pfeifer U (1987) Short-term stimulation by propranolol and verapamil of cardiac cellular autophagy. J Mol Cell Cardiol 19:1169–1178
Buck ML (2010) Oral propranolol for hemangiomas of infancy. Pediatric Pharmacotherapy 16(8)
Liu K, Liu C, Shen L, Shi J, Zhang T, Zhou Y, Zhou L, Sun X (2011) Therapeutic effects of rapamycin on MPTP-induced Parkinsonism in mice. Neurochem Int. doi:10.1016/j.neuint.2011.05.011
Morselli E, Galluzzi L, Kepp O, Criollo A, Maiuri MC, Tavernarakis N, Madeo F, Kroemer G (2009) Autophagy mediates pharmacological lifespan extension by spermidine and resveratrol. Aging (Albany NY) 1:961–970
Konrady AO, Kasherininov YR, Shavarov AA, Shavarova EK, Vachrameeva NV, Krutikov AN, Smirnova EV, Shlyakhto EV (2006) How can we block sympathetic overactivity? Effects of rilmenidine and atenolol in overweight hypertensive patients. J Hum Hypertens 20:398–406. doi:10.1038/sj.jhh.1002004
McLean K, VanDeVen NA, Sorenson DR, Daudi S, Liu JR (2009) The HIV protease inhibitor saquinavir induces endoplasmic reticulum stress, autophagy, and apoptosis in ovarian cancer cells. Gynecol Oncol 112:623–630
Zorrilla CD, Van Dyke R, Bardeguez A, Acosta EP, Smith B, Hughes MD, Huang S, Watts DH, Heckman B, Jimenez E et al (2007) Clinical response and tolerability to and safety of saquinavir with low-dose ritonavir in human immunodeficiency virus type 1-infected mothers and their infants. Antimicrob Agents Chemother 51:2208–2210
Jiang Q, Li F, Shi K, Yang Y, Xu C (2012) Sodium selenite-induced activation of DAPK promotes autophagy in human leukemia HL60 cells. BMB Rep 45:194–199
Bareford MD, Hamed HA, Tang Y, Cruickshanks N, Burow ME, Fisher PB, Moran RG, Nephew KP, Grant S, Dent P (2011) Sorafenib enhances pemetrexed cytotoxicity through an autophagy-dependent mechanism in cancer cells. Autophagy 7:1261–1262
Lam ET, Ringel MD, Kloos RT, Prior TW, Knopp MV, Liang J, Sammet S, Hall NC, Wakely PE Jr, Vasko VV et al (2010) Phase II clinical trial of sorafenib in metastatic medullary thyroid cancer. J Clin Oncol 28:2323–2330
Tirupathi Pichiah PB, Suriyakalaa U, Kamalakkannan S, Kokilavani P, Kalaiselvi S, SankarGanesh D, Gowri J, Archunan G, Cha YS, Achiraman S (2011) Spermidine may decrease ER stress in pancreatic beta cells and may reduce apoptosis via activating AMPK dependent autophagy pathway. Med Hypotheses 77:677–679
Jiang J, Maeda A, Ji J, Baty CJ, Watkins SC, Greenberger JS, Kagan VE (2011) Are mitochondrial reactive oxygen species required for autophagy? Biochem Biophys Res Commun 412:55–60
Chen Y, Azad MB, Gibson SB (2009) Superoxide is the major reactive oxygen species regulating autophagy. Cell Death Differ 16:1040–1052
Bursch W, Ellinger A, Kienzl H, Torok L, Pandey S, Sikorska M, Walker R, Hermann RS (1996) Active cell death induced by the anti-estrogens tamoxifen and ICI 164 384 in human mammary carcinoma cells (MCF-7) in culture: the role of autophagy. Carcinogenesis 17:1595–1607
Lawrence JA, Adamson PC, Caruso R, Chow C, Kleiner D, Murphy RF, Venzon DJ, Shovlin M, Noone M, Merino M et al (2001) Phase I clinical trial of alitretinoin and tamoxifen in breast cancer patients: toxicity, pharmacokinetic, and biomarker evaluations. J Clin Oncol 19:2754–2763
Fujisaka Y, Yamada Y, Yamamoto N, Horiike A, Tamura T (2010) A Phase 1 clinical study of temsirolimus (CCI-779) in Japanese patients with advanced solid tumors. Jpn J Clin Oncol 40:732–738
Zeng X, Kinsella TJ (2010) BNIP3 is essential for mediating 6-thioguanine- and 5-fluorouracil-induced autophagy following DNA mismatch repair processing. Cell Res 20:665–675
Petit E, Langouet S, Akhdar H, Nicolas-Nicolaz C, Guillouzo A, Morel F (2008) Differential toxic effects of azathioprine, 6-mercaptopurine and 6-thioguanine on human hepatocytes. Toxicol in Vitro 22:632–642
Badham JN, Bardon LM, Reeves PO, Young AM (1963) A trial of thioridazine in mental deficiency. Br J Psychiatry 109:408–410
Kruger U, Wang Y, Kumar S, Mandelkow EM (2011) Autophagic degradation of tau in primary neurons and its enhancement by trehalose. Neurobiol Aging 33(10):2291–2305
Khunger N, Sarkar R, Jain RK (2004) Tretinoin peels versus glycolic acid peels in the treatment of melasma in dark-skinned patients. Dermatol Surg 30:756–760, discussion 760
Takeuchi H, Kondo Y, Fujiwara K, Kanzawa T, Aoki H, Mills GB, Kondo S (2005) Synergistic augmentation of rapamycin-induced autophagy in malignant glioma cells by phosphatidylinositol 3-kinase/protein kinase B inhibitors. Cancer Res 65:3336–3346
Klein B, Worndl K, Lutz-Meindl U, Kerschbaum HH (2011) Perturbation of intracellular K(+) homeostasis with valinomycin promotes cell death by mitochondrial swelling and autophagic processes. Apoptosis 16:1101–1117
Duenas-Gonzalez A, Candelaria M, Perez-Plascencia C, Perez-Cardenas E, de la Cruz-Hernandez E, Herrera LA (2008) Valproic acid as epigenetic cancer drug: preclinical, clinical and transcriptional effects on solid tumors. Cancer Treat Rev 34:206–222
Pennock GD, Dalton WS, Roeske WR, Appleton CP, Mosley K, Plezia P, Miller TP, Salmon SE (1991) Systemic toxic effects associated with high-dose verapamil infusion and chemotherapy administration. J Natl Cancer Inst 83:105–110
Claerhout S, Lim JY, Choi W, Park YY, Kim K, Kim SB, Lee JS, Mills GB, Cho JY (2011) Gene expression signature analysis identifies vorinostat as a candidate therapy for gastric cancer. PLoS One 6:e24662
Ramalingam SS, Kummar S, Sarantopoulos J, Shibata S, LoRusso P, Yerk M, Holleran J, Lin Y, Beumer JH, Harvey RD et al (2010) Phase I study of vorinostat in patients with advanced solid tumors and hepatic dysfunction: a National Cancer Institute Organ Dysfunction Working Group study. J Clin Oncol 28:4507–4512
Yang D, Liu H, Goga A, Kim S, Yuneva M, Bishop JM (2010) Therapeutic potential of a synthetic lethal interaction between the MYC proto-oncogene and inhibition of aurora-B kinase. Proc Natl Acad Sci U S A 107:13836–13841
Bauer PO, Wong HK, Oyama F, Goswami A, Okuno M, Kino Y, Miyazaki H, Nukina N (2009) Inhibition of Rho kinases enhances the degradation of mutant huntingtin. J Biol Chem 284:13153–13164
Chen SY, Chiu LY, Maa MC, Wang JS, Chien CL, Lin WW (2011) zVAD-induced autophagic cell death requires c-Src-dependent ERK and JNK activation and reactive oxygen species generation. Autophagy 7:217–228
Ye YC, Yu L, Wang HJ, Tashiro S, Onodera S, Ikejima T (2011) TNFalpha-induced necroptosis and autophagy via supression of the p38-NF-kappa β survival pathway in L929 cells. J Pharmacol Sci 117:160–169
Major P, Lortholary A, Hon J, Abdi E, Mills G, Menssen HD, Yunus F, Bell R, Body J, Quebe-Fehling E et al (2001) Zoledronic acid is superior to pamidronate in the treatment of hypercalcemia of malignancy: a pooled analysis of two randomized, controlled clinical trials. J Clin Oncol 19:558–567
Shen S, Kepp O, Michaud M, Martins I, Minoux H, Metivier D, Maiuri MC, Kroemer RT, Kroemer G (2011) Association and dissociation of autophagy, apoptosis and necrosis by systematic chemical study. Oncogene 30:4544–4556
Hill JA, O’Brien JT, Alpert JS, Gore JM, Zusman RM, Christensen D, Boucher CA, Vetrovec G, Borer JS, Friedman C et al (1985) Effect of bepridil in patients with chronic stable angina: results of a multicenter trial. Circulation 71:98–103
Lai JH, Ho LJ, Kwan CY, Chang DM, Lee TC (1999) Plant alkaloid tetrandrine and its analog block CD28-costimulated activities of human peripheral blood T cells: potential immunosuppressants in transplantation immunology. Transplantation 68:1383–1392
Sharpe DS (1962) A controlled trial of trifluoperazine in the treatment of the mentally subnormal patient. J Ment Sci 108:220–224
Lyko F, Brown R (2005) DNA methyltransferase inhibitors and the development of epigenetic cancer therapies. J Natl Cancer Inst 97:1498–1506
Kaminskas E, Farrell A, Abraham S, Baird A, Hsieh LS, Lee SL, Leighton JK, Patel H, Rahman A, Sridhara R et al (2005) Approval summary: azacitidine for treatment of myelodysplastic syndrome subtypes. Clin Cancer Res 11:3604–3608
Cashen AF, Schiller GJ, O’Donnell MR, DiPersio JF (2010) Multicenter, phase II study of decitabine for the first-line treatment of older patients with acute myeloid leukemia. J Clin Oncol 28:556–561
Merskey H (1958) A clinical and psychometric study of the effects of procaine amide in Huntington’s chorea. J Ment Sci 104:411–420
Mereles D, Hunstein W (2011) Epigallocatechin-3-gallate (EGCG) for clinical trials: more pitfalls than promises? Int J Mol Sci 12:5592–5603
Singh N, Duenas-Gonzalez A, Lyko F, Medina-Franco JL (2009) Molecular modeling and molecular dynamics studies of hydralazine with human DNA methyltransferase 1. ChemMedChem 4:792–799
Stiell IG, Clement CM, Symington C, Perry JJ, Vaillancourt C, Wells GA (2007) Emergency department use of intravenous procainamide for patients with acute atrial fibrillation or flutter. Acad Emerg Med 14:1158–1164
Liu Z, Liu S, Xie Z, Pavlovicz RE, Wu J, Chen P, Aimiuwu J, Pang J, Bhasin D, Neviani P et al (2009) Modulation of DNA methylation by a sesquiterpene lactone parthenolide. J Pharmacol Exp Ther 329:505–514
Curry EA 3rd, Murry DJ, Yoder C, Fife K, Armstrong V, Nakshatri H, O’Connell M, Sweeney CJ (2004) Phase I dose escalation trial of feverfew with standardized doses of parthenolide in patients with cancer. Investig New Drugs 22:299–305
Liu Z, Xie Z, Jones W, Pavlovicz RE, Liu S, Yu J, Li PK, Lin J, Fuchs JR, Marcucci G et al (2009) Curcumin is a potent DNA hypomethylation agent. Bioorg Med Chem Lett 19:706–709
Hsu CH, Cheng AL (2007) Clinical studies with curcumin. Adv Exp Med Biol 595:471–480
Lin RK, Hsu CH, Wang YC (2007) Mithramycin A inhibits DNA methyltransferase and metastasis potential of lung cancer cells. Anticancer Drugs 18:1157–1164
Liacini A, Sylvester J, Li WQ, Zafarullah M (2005) Mithramycin downregulates proinflammatory cytokine-induced matrix metalloproteinase gene expression in articular chondrocytes. Arthritis Res Ther 7:R777–R783
Kuck D, Singh N, Lyko F, Medina-Franco JL (2010) Novel and selective DNA methyltransferase inhibitors: docking-based virtual screening and experimental evaluation. Bioorg Med Chem 18:822–829
Kuck D, Caulfield T, Lyko F, Medina-Franco JL (2010) Nanaomycin A selectively inhibits DNMT3B and reactivates silenced tumor suppressor genes in human cancer cells. Mol Cancer Ther 9:3015–3023
Acknowledgments
The authors would like to acknowledge Dr. Annalise M. Nawrocki for comments on an earlier version of this manuscript. Generous support for this work was provided by the Retina Macular Degeneration Fund, an AF-IDA grant from UCD and NIH 1R01EY021024-01A2 to ZSM. The work is supported by Grant of Presidium of RAS “Structure-functional organization of chromosomes in cell cycle (12-С-4-1007)” to AAM.
Open Access
This article is distributed under the terms of the Creative Commons Attribution License which permits any use, distribution, and reproduction in any medium, provided the original author(s) and the source are credited.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 2.0 International License (https://creativecommons.org/licenses/by/2.0), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
About this article
Cite this article
Zhavoronkov, A., Smit-McBride, Z., Guinan, K.J. et al. Potential therapeutic approaches for modulating expression and accumulation of defective lamin A in laminopathies and age-related diseases. J Mol Med 90, 1361–1389 (2012). https://doi.org/10.1007/s00109-012-0962-4
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00109-012-0962-4