Abstract
Purpose
The construction of Technosols is an emergent technology based on the assemblage of technogenic materials for ecological reclamation of polluted land and waste recycling. Although this technology is in expansion, knowledge about the microbial communities in Technosols is limited, despite their central role in ecosystem functioning. In this 2-year study, the diversity and the abundance of total and functional microbial communities were characterized in two types of Technosols constructed to reclaim contaminated sites.
Materials and methods
The structure of the microbial community was analyzed by automated ribosomal intergenic spacer analysis fingerprinting in both types of Technosols, and the taxonomic diversity was further assessed by 16S rRNA clone library sequencing. Real-time PCR was used to quantify the abundance of the total bacterial and crenarchaeal community and of the functional guilds involved in N-cycling.
Results and discussion
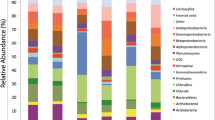
16S rRNA sequencing showed that Proteobacteria was the main phylum in the Technosols (50–80 %). The other significant phyla identified were Bacteroidetes, Firmicutes, Choloroflexi, and Actinobacteria. Real-time PCR quantification of the abundance of ammonia oxidizers, nitrate reducing, and denitrifying microbial communities involved in nitrogen cycling revealed that bacterial ammonia oxidizers were more abundant than crenarchaeal ammonia oxidizers. A high spatial variability of the microbial community, which decreased with time, was also observed.
Conclusions
At the phyla and class levels, the composition of the microbial community in constructed Technosols was similar to “natural” soils. Both the total bacteria and microbial guilds involved in N-cycling were abundant, but in contrast to most natural soils, bacteria and not crenarchaea were the numerically dominant ammonia oxidizers in both types of Technosols. The decrease with time of the variability in microbial community structure support early pedogenic evolution of recently constructed Technosols.





Similar content being viewed by others
References
Addinsoft SARL (2009) XLSTAT software. (ed) Paris, France
AFNOR (Association Française de Normalisation) (2004) Evaluation de la qualité des sols volume 1: méthodes d’analyse chimique. La Plaine Saint-Denis Cedex, 461 pp
Basol Database (2011) Pollution des sols; Ministère de l’Écologie, de l’Énergie, du Développement Durable et de la Mer. http://basol.environnement.gouv.fr Accessed 26 April 2012
Bodelier P (2011) Towards understanding, managing and protecting microbial ecosystems. Front Microbiol 2:80
Bru D, Sarr A, Philippot L (2007) Relative abundances of proteobacterial membrane-bound and periplasmic nitrate reductases in selected environments. Appl Environ Microbiol 73:5971–5974
Bru D, Ramette A, Saby NPA, Dequiedt S, Ranjard L, Jolivet C, Arrouays D, Philippot L (2011) Determinants of the distribution of nitrogen-cycling microbial communities at the landscape scale. ISME J 5:532–542
COM (2006) Directive of the European parliament and of the council establishing a framework for the protection of soil and amending Directive 2004/35/EC. Commission of the European community. Brussels. http://eur-lex.europa.eu/LexUriServ/LexUriServ.do?uri=COM:2006:0232:FIN:EN:PDF Accessed 26 April 2012
Conrad R (1996) Soil microorganisms as controllers of atmospheric trace gases (H2, CO, CH4, OCS, N2O, and NO). Microbiol Rev 60:609–640
Erguder TH, Boon N, Wittebolle L, Marzorati M, Verstraete W (2009) Environmental factors shaping the ecological niches of ammonia-oxidizing Archaea. FEMS Microbiol Rev 33:855–869
Haritash AK, Kaushik CP (2009) Biodegradation aspects of polycyclic aromatic hydrocarbons (PAHs): a review. J Hazard Mater 169:1–15
Henry S, Baudoin E, Lopez-Gutierrez JC, Martin-Laurent F, Baumann A, Philippot L (2004) Quantification of denitrifying bacteria in soils by nirK gene targeted real-time PCR. J Microbiol Methods 59:327–335
Henry S, Bru D, Stres B, Hallet S, Philippot L (2006) Quantitative detection of the nosZ gene, encoding nitrous oxide reductase, and comparison of the abundances of 16S rRNA, narG, nirK, and nosZ genes in soils. Appl Environ Microbiol 72:5181–5189
Huber T, Faulkner G, Hugenholtz P (2004) Bellerophon: a program to detect chimeric sequences in multiple sequence alignments. Bioinformatics 20:2317–2319
Janssen PH (2006) Identifying the dominant soil bacterial taxa in libraries of 16S rRNA and 16S rRNA genes. Appl Environ Microbiol 72:1719–1728
Jones CM, Stres B, Rosenquist M, Hallin S (2008) Phylogenetic analysis of nitrite, nitric oxide, and nitrous oxide respiratory enzymes reveal a complex evolutionary history for denitrification. Mol Biol Evol 25:1955–1966
Leininger S, Urich T, Schloter M, Schwark L, Qi J, Nicol GW, Prosser JI, Schuster SC, Schleper C (2006) Archaea predominate among ammonia-oxidizing prokaryotes in soils. Nature 442:806–809
Lopez-Gutierrez JC, Henry S, Hallet S, Martin-Laurent F, Catroux G, Philippot L (2004) Quantification of a novel group of nitrate-reducing bacteria in the environment by real-time PCR. J Microbiol Methods 57:399–407
Lozupone C, Knight R (2005) UniFrac: a new phylogenetic method for comparing microbial communities. Appl Environ Microbiol 71:8228–8235
Martin-Laurent F, Philippot L, Hallet S, Chaussod R, Germon JC, Soulas G, Catroux G (2001) DNA extraction from soils: old bias for new microbial diversity analysis methods. Appl Environ Microbiol 67:2354–2359
Mertens J, Broos K, Wakelin SA, Kowalchuk GA, Springael D, Smolders E (2009) Bacteria, not Archaea, restore nitrification in a zinc-contaminated soil. ISME J 3:916–923
Nannipieri P, Ascher J, Ceccherini MT, Landi L, Pietramellara G, Renella G (2003) Microbial diversity and soil functions. Eur J Soil Sci 54:655–670
Nemergut DR, Cleveland CC, Wieder WR, Washenberger CL, Townsend AR (2010) Plot-scale manipulations of organic matter inputs to soils correlate with shifts in microbial community composition in a lowland tropical rain forest. Soil Biol Biochem 42:2153–2160
Ochsenreiter T, Selezi D, Quaiser A, Bonch-Osmolovskaya L, Schleper C (2003) Diversity and abundance of Crenarchaeota in terrestrial habitats studied by 16S RNA surveys and real time PCR. Environ Microbiol 5:787–797
Petric I, Philippot L, Abbate C, Bispo A, Chesnot T, Hallin S, Laval K, Lebeau T, Lemanceau P, Leyval C, Lindström K, Pandard P, Romero E, Sarr A, Schloter M, Simonet P, Smalla K, Wilke BM, Martin-Laurent F (2011) Inter-laboratory evaluation of the ISO standard 11063 “soil quality—method to directly extract DNA from soil samples”. J Microbiol Methods 84:454–460
Petrić I, Hršak D, Fingler S, Udiković-Kolić N, Bru D, Martin-Laurent F (2011) Insight in the PCB-degrading functional community in long-term contaminated soil under bioremediation. J Soils Sediments 11:290–300
Philippot L, Cuhel J, Saby NPA, Cheneby D, Chronakova A, Bru D, Arrouays D, Martin-Laurent F, Simek M (2009) Mapping field-scale spatial patterns of size and activity of the denitrifier community. Environ Microbiol 11:1518–1526
Philippot L, Abbate C, Bispo A, Chesnot T, Hallin S, Lemanceau P, Lindström K, Pandard P, Romero E, Schloter M, Simonet P, Smalla K, Wilke B-M, Petric I, Martin-Laurent F (2010) Soil microbial diversity: an ISO standard for soil DNA extraction. J Soils Sediments 10:1344–1345
Philippot L, Andert J, Jones CM, Bru D, Hallin S (2011) Importance of denitrifiers lacking the genes encoding the nitrous oxide reductase for N2O emissions from soil. Glob Chang Biol 17:1497–1504
Prosser JI, Nicol GW (2008) Relative contributions of Archaea and Bacteria to aerobic ammonia oxidation in the environment. Environ Microbiol 10:2931–2941
Ranjard L, Poly F, Combrisson J, Richaume A, Gourbiere F, Thioulouse J, Nazaret S (2000) Heterogeneous cell density and genetic structure of bacterial pools associated with various soil microenvironments as determined by enumeration and DNA fingerprinting approach (RISA). Microb Ecol 39:263–272
Schleper C, Nicol GW (2010) Ammonia-oxidising Archaea—physiology, ecology and evolution. Adv Microb Physiol 57:1–41
Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB, Lesniewski RA, Oakley BB, Parks DH, Robinson CJ, Sahl JW, Stres B, Thallinger GG, Van Horn DJ, Weber CF (2009) Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol 75:7537–7541
Sere G, Schwartz C, Ouvrard S, Sauvage C, Renat JC, Morel JL (2008) Soil construction: a step for ecological reclamation of derelict lands. J Soils Sediments 8:130–136
Sere G, Schwartz C, Ouvrard S, Renat JC, Watteau F, Villemin G, Morel JL (2010) Early pedogenic evolution of constructed Technosols. J Soils Sediments 10:1246–1254
Throbäck IN, Enwall K, Jarvis Å, Hallin S (2004) Reassessing PCR primers targeting nirS, nirK and nosZ genes for community surveys of denitrifying bacteria with DGGE. FEMS Microbiol Ecol 49:401–417
Torsvik V, Øvreås L (2002) Microbial diversity and function in soil: from genes to ecosystems. Curr Opin Microbiol 5:240–245
Tourna M, Freitag TE, Nicol GW, Prosser JI (2008) Growth, activity and temperature responses of ammonia-oxidizing Archaea and Bacteria in soil microcosms. Environ Microbiol 10:1357–1364
van der Heijden MGA, Bardgett RD, van Straalen NM (2008) The unseen majority: soil microbes as drivers of plant diversity and productivity in terrestrial ecosystems. Ecol Lett 11:296–310
Vinas M, Sabate J, Espuny MJ, Solanas AM (2005) Bacterial community dynamics and polycyclic aromatic hydrocarbon degradation during bioremediation of heavily creosote-contaminated soil. Appl Environ Microbiol 71:7008–7018
Wardle DA, Giller KE (1996) The quest for a contemporary ecological dimension to soil biology—discussion. Soil Biol Biochem 28:1549–1554
Watanabe K, Kodama Y, Harayama S (2001) Design and evaluation of PCR primers to amplify bacterial 16S ribosomal DNA fragments used for community fingerprinting. J Microbiol Methods 44:253–262
Wessen E, Soderstrom M, Stenberg M, Bru D, Hellman M, Welsh A, Thomsen F, Klemedtson L, Philippot L, Hallin S (2011) Spatial distribution of ammonia-oxidizing bacteria and Archaea across a 44-hectare farm related to ecosystem functioning. ISME J 5:1213–1225
Whitman WB, Coleman DC, Wiebe WJ (1998) Prokaryotes: the unseen majority. Proc Natl Acad Sci U S A 95:6578–6583
WRB IWG (2006) World reference base for soil resource. World resources Report 103, FAO, Rome, 2006 (2nd ed) (2006): World reference base for soil resource. World resources Report 103, FAO, Rome
Zhang LM, Offre PR, He JZ, Verhamme DT, Nicol GW, Prosser JI (2010) Autotrophic ammonia oxidation by soil thaumarchaea. Proc Natl Acad Sci U S A 107:17240–17245
Acknowledgments
The PhD work of Farhan Hafeez was funded by the Higher Education Commission (Pakistan) and supported by the Doctoral School E2S of the University of Burgundy (France). This work was part of the “Biotechnosol program” (contracts no. S.6-0006653 with INPL and S.6-0006654 with INRA) coordinated by Dr J Cortet and carried out as part of the Gessol program “Fonctions environnementales des sols et gestion du patrimoine sol” funded by the French Ministry of Ecology in cooperation with the ADEME. We should also like to thank the Valterra Company (Jean-Christophe RENAT and Sophie GUIMONT) for the soil construction and the technical staff of the Laboratoire Sols et Environnement, including Jean-Claude Bégin, Adeline Bouchard, Stéphane Colin, Romain Goudon, and Alain Rakoto, as well as the GISFI, particularly Noele Raoult, Cindy Messana, and Lucas Charrois, for technical assistance and management of field plots.
Author information
Authors and Affiliations
Corresponding author
Additional information
Responsible editor: Yanfen Wang
Rights and permissions
About this article
Cite this article
Hafeez, F., Martin-Laurent, F., Béguet, J. et al. Taxonomic and functional characterization of microbial communities in Technosols constructed for remediation of a contaminated industrial wasteland. J Soils Sediments 12, 1396–1406 (2012). https://doi.org/10.1007/s11368-012-0563-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11368-012-0563-4