Abstract
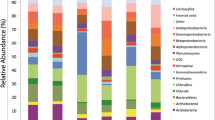
The Cerrado biome in the Sete Cidades National Park, an Ecological Reserve in Northeastern Brazil, has conserved its native biodiversity and presents a variety of plants found in other savannas in Brazil. Despite this finding the soil microbial diversity and community structure are poorly understood. Therefore, we described soil bacterial diversity and distribution along a savanna vegetation gradient taking into account the prevailing environmental factors. The bacterial composition was retrieved by sequencing a fragment of the 16S ribosomal RNA gene. The bacterial operational taxonomic units (OTUs) were assigned to 37 different phyla, 96 classes, and 83 genera. At the phylum level, a core comprised by Proteobacteria, Acidobacteria, Actinobacteria, Firmicutes, Verrucomicrobia and Planctomycetes, was detected in all areas of Cerrado. ‘Cerrado stricto sensu’ and ‘Cerradao’ share more similarities between edaphic properties and vegetation and also present more similar bacterial communities, while ‘Floresta decidual’ and ‘Campo graminoide’ show the largest environmental differences and also more distinct bacterial communities. Proteobacteria (26%), Acidobacteria (21%) and Actinobacteria (21%) were the most abundant phyla within the four areas. All the samples present similar bacteria richness (alpha diversity) and the observed differences among them (beta diversity) were more related to the abundance of specific taxon OTUs compared to their presence or absence. Total organic C, N and P are the main abiotic factors structuring the bacterial communities. In summary, our findings show the bacterial community structure was clearly different across the Cerrado gradient, but that these environments share a bacterial phylum-core comprising Proteobacteria, Acidobacteria, Actinobacteria, Verrucomicrobia and Planctomycetes with other Brazilian savannas.






Similar content being viewed by others
References
Araujo JF, Castro AP, Costa MMC, Togawa RC, Pappas Júnior GJ, Quirino BF, Bustamante MMC, Williamson L, Handelsman J, Krüger RH (2012) Characterization of soil bacterial assemblies in Brazilian savanna-like vegetation reveals acidobacteria dominance. Microb Ecol 64:760–770
Bardgett RD, Shine A (1999) Linkages between litter diversity, soil microbial biomass and ecosystem function in temperate grasslands. Soil Biol Biochem 31:317–321
Bergmann GT, Bates ST, Eilers KG, Lauber CL, Caporaso JG, Walters WA, Knight R, Fierer N (2011) The under-recognized dominance of verrucomicrobia in soil bacterial communities. Soil Biol Biochem 43:1450–1455
Berry AM, Harriott OT, Moreau RA, Osman SF, Benson DR, Jones AD (1993) Hopanoid lipids compose the Frankia vesicle envelope, presumptive barrier of oxygen diffusion to nitrogenase. Proc Natl Acad Sci USA 90:6091–6094
Bresolin JD, Bustamante MMC, Krüger RH, Silva MRSS, Perez KS (2010) Structure and composition of bacterial and fungal community in soil under soybean monoculture in the Brazilian Cerrado. Braz J Microbiol 41:391–403
Bru D, Ramette A, Saby NP, Dequiedt S, Ranjard L, Jolivet C, Arrouays D, Philippot L (2011) Determinants of the distribution of nitrogen-cycling microbial communities at the landscape scale. ISME J 5:532–542
Caporaso JG, Bittinger K, Bushman FD, DeSantis TZ, Andersen GL, Knight R (2010a) PyNAST: a flexible tool for aligning sequences to a template alignment. Bioinformatics 26:266–267
Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Peña AG, Goodrich JK, Gordon JI, Huttley GA, Kelley ST, Knights D, Koenig JE, Ley RE, Lozupone CA, McDonald D, Muegge BD, Pirrung M, Reeder J, Sevinsky JR, Turnbaugh PJ, Walters WA, Widmann J, Yatsunenko T, Zaneveld J, Knight R (2010b) QIIME allows analysis of high-throughput community sequencing data. Nat Methods 7:335–336
Caporaso JG, Lauber CL, Walters WA, Berg-Lyons D, Lozupone CA, Turnbaugh PJ, Fierer N, Knight R (2011) Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. Proc Natl Acad Sci USA 108:4516–4522
Cassman NA, Leite MFA, Pan Y, de Hollander M, van Veen JA, Kuramae EE (2016) Plant and soil fungal but not soil bacterial communities are linked in long-term fertilized grassland. Sci Rep 6:23680
Castro AAJF, Martins FR, Fernandes AG (1998) The woody flora of Cerrado vegetation in the state of Piauí, Northeastern Brazil. Edinb J Botany 55:455–472
Castro AAJF, Martins FR, Tamashiro JY, Shepherd GJ (1999) How rich is the flora of Brazilian Cerrados? Ann Missouri Bot Gard 86:192–224
Castro AP, Silva MRSS, Quirino BF, Bustamante MMC, Krüger RH (2016) Microbial diversity in Cerrado biome (Neotropical Savanna) soils. PLoS ONE 11:e0148785
Coutinho LM (1978) O conceito de cerrado. Rev Bras Bot 1:17–23
DeMandal S, Zothansanga AKP, Bisht SS, Kumar NS (2015) First report of bacterial community from a bat guano using Illumina next-generation sequencing. Gen Data 4:99–101
DeSantis TZ, Hugenholtz P, Larsen N, Rojas M, Brodie EL, Keller K, Huber T, Dalevi D, Hu P, Andersen GL (2006) Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. App Environ Microbiol 72:5069–5072
Edgar RC (2013) UPARSE: highly accurate OTU sequences from microbial amplicon reads. Nat Methods 10:996–998
Edgar RC, Haas BJ, Clemente JC, Quince C, Knight R (2011) UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 27:2194–2200
Forzza RC, Leitman PM, Costa A, Carvalho AA, Peixoto AL, Walter BMT, Bicudo C, Zappi D, Costa DP, Lleras E, Martinelli G, Lima HC, Prado J, Stehmann JR, Baumgratz JFA, Pirani JR, Sylvestre LS, Maia LC, Lohmann LG, Paganucci L, Silveira M, Nadruz M, Mamede MCH, Bastos MNC, Morim MP, Barbosa MR, Menezes M, Hopkins M, Secco R, Cavalcanti T, Souza VC (2010) Catalogo de plantas e fungos do Brasil. Instituto de Pesquisas Jardim Botanico, Rio de Janeiro
Gupta RS (2004) The phylogeny and signature sequences characteristics of fibrobacteres, chlorobi, and bacteroidetes. Critical Rev Microbiol 30:123–140
Hartmann M, Niklaus PA, Zimmermann S (2014) Resistance and resilience of the forest soil microbiome to logging-associated compaction. ISME J 8:226–244
IBDF (1979) Plano de manejo: parque nacional de sete cidades. Fundação Brasileira para a Conservação da Natureza, Brasília
Illumina (2013) 16S metagenomic sequencing library preparation. Illumina Guides http://support.illumina.com/downloads/16s_metagenomic_sequencing_library_preparation.html. Accessed 6 Mar 2014
Janssen PH (2006) Identifying the dominant soil bacterial taxa in libraries of 16S rRNA and 16S rRNA genes. Appl Environ Microbiol 73:1719–1728
Kannenberg EL, Perzi M, Muller P, Hartner T, Poralla K (1996) Hopanoid lipids in Bradyrhizobium and other plant-associated bacteria and cloning of the Bradyrhizobium squalene-hopene cyclase. Pl Soil 186:107–112
Kopecky J, Kyselkova M, Omelka M, Cermak L, Novotna J, Grundmann GL, Moenne-Loccoz Y, Sagova-Mareckova M (2011) Actinobacterial community dominated by a distinct clade in acidic soil of a waterlogged deciduous forest. FEMS Microbiol Ecol 78:386–394
Lodha TD, Srinivas A, Sasikala C, Ramana CV (2015) Hopanoid inventory of Rhodoplanes spp. Arch Microbiol 197:861–867
Lozupone C, Knight R (2005) UniFrac: a new phylogenetic method for comparing microbial communities. Appl Environ Microbiol 71:8228–8235
Martiny JB, Eisen JA, Penn K, Allison SD, Horner-Devine MC (2011) Drivers of bacterial beta-diversity depend on spatial scale. Proc Natl Acad Sci 108:7850–7854
Mishra S, Lee WA, Hooijer A, Reuben S, Sudiana IM, Idris A, Swarup S (2014) Microbial and metabolic profiling reveal strong influence of water table and land-use patterns on classification of degraded tropical peatlands. Biogeosciences 11:1727–1741
Monciardini P, Cavaletti L, Schumann P, Rohde M, Donadio S (2003) Conexibacter woesei gen. nov., sp. nov., a novel representative of a deep evolutionary line of descent within the class Actinobacteria. Int J Syst Evol Microbiol 53:569–576
Mota FF, Gomes EA, Marriel IE, Paiva E, Seldin L (2008) Tolerant and aluminum-sensitive maize (Zea mays L.) lines cultivated in unlimed and limed Cerrado soil. Microbiol Biotechnol 18:805–814
Myers N, Mittermeier RA, Mittermeier CG, Fonseca GAB, Kent J (2000) Biodiversity. hotspots for conservation priorities. Nature 403:853–858
Okamura K, Kanbe T, Hiraishi A (2009) Rhodoplanes serenus sp. nov., a purple non-sulfur bacterium isolated from pond water. Int J Syst Evol Microbiol 59:531–535
Oksanen J, Blanchet FG, Kindt R, Legendre P, Minchin PR, O’Hara RB, Simpson GL, Solymos P, Stevens MHH, Wagner H (2015) Vegan: community ecology package. R package version 2.2–1. http://cran.r-project.org/package=vegan
Oliveira MEA, Martins FR, Castro AAJF, Santos JR (2007) Classes de cobertura vegetal do parque nacional de sete cidades (transição campo-floresta) utilizando imagens TM/Landsat, NE do Brasil. Proceedings XIII Simpósio Brasileiro de Sensoriamento Remoto Anais 13:1775–1783
Peixoto RS, Coutinho HLC, Madari B, Machado PLOA, Rumjanek NG, Van Elsas JD, Seldin L, Rosado AS (2006) Soil aggregation and bacterial community structure as affected by tillage and cover cropping in the Brazilian Cerrados. Soil Till Res 90:16–28
Pindi PK, Sultana T, Vootla PK (2014) Plant growth regulation of Bt-cotton through Bacillus species. 3 Biotech 4:305–315
Pitombo LM, Carmo JB, Hollander M, Rossetto R, López MV, Cantarella H, Kuramae EE (2015) Exploring soil microbial 16S rRNA sequence data to increase carbon yield and nitrogen efficiency of a bioenergy crop. GCB Bioenergy. doi:10.1111/gcbb.12284
Pylro VS, Roesch LFW, Ortega JM, Amaral AM, Tótola MR, Hirsch PR, Rosado AS, Góes-Neto A, Silva ALC, Rosa CA, Morais DK, Andreote FD, Duarte GF, Melo IS, Seldin L, Lambais MR, Hungria M, Peixoto RS, Kruger RH, Tsai SM, Azevedo V (2014) Brazilian microbiome project: revealing the unexplored microbial diversity–challenges and prospects. Microb Ecol 67:237–241
Pylro VS, Morais DK, Oliveira FS, Santos FG, Lemos LN, Oliveira G, Roesch LFW (2016) BMPOS: a flexible and user-friendly tool sets for microbiome studies. Microb Ecol 73:443–447
Quirino BF, Pappas GJ, Tagliaferro AC, Collevatti RG, Neto EL, Silva MR, Bustamante MM, Kruger RH (2009) Molecular phylogenetic diversity of bacteria associated with soil of savanna-like Cerrado vegetation. Microbiol Res 164:59–70
Rachid CTCC, Santos AL, Piccolo MC, Balieiro FC, Coutinho HLC, Peixoto RS (2013) Effect of sugarcane burning or green harvest methods on the Brazilian Cerrado soil bacterial community structure. PLoS ONE 8:e59342
Rampelotto PH, Ferreira AS, Barboza AD, Roesch LF (2013) Changes in diversity, abundance, and structure of soil bacterial communities in Brazilian Savanna under different land use systems. Microb Ecol 66:593–607
Rodrigues JLM, Pellizari VH, Mueller R, Baek K, Jesus EC, Paula FS, Mirza B, Hamaoui GS, Tsai SM, Feigl B, Tiedje JM, Bohannan JM, Nüsslein K (2013) Conversion of the Amazon rainforest to agriculture results in biotic homogenization of soil bacterial communities. Proc Natl Acad Sci USA 110:988–993
Rysanek D, Hrckova K, Skaloud P (2015) Global ubiquity and local endemism of free-living terrestrial protists: phylogeographic assessment of the streptophyte alga Klebsormidium. Environ Microbiol 17:689–698
Steenbergh AK, Bodelier PLE, Hoogveld HL, Slomp CP, Laanbroek HJ (2015) Phylogenetic characterization of phosphatase-expressing bacterial communities in baltic sea sediments. Microbes Environ 30:192–195
Tedesco MJ, Gianello C, Bissani CA, Bohnen H, Volkweiss SJ (1995) Análise de solo, plantas e outros materiais. Universidade Federal do Rio Grande do Sul, Porto Alegre, p 147
Vandekerckhove TTM, Willems A, Gillis M, Coomans A (2000) Occurrence of novel verrucomicrobial species, endosymbiotic and associated with parthenogenesis in Xiphinema americanum-group species (Nematoda, Longidoridae). Int J Syst Evol Microbiol 50:2197–2205
Wyrick P (2000) Intracellular survival by Chlamydia. Cell Microbiol 2:275–282
Xiong J, Liu Y, Lin X, Zhang H, Zeng J, Hou J, Yang Y, Yao T, Knight R, Chu H (2012) Geographic distance and pH drive bacterial distribution in alkaline lake sediments across Tibetan Plateau. Environ Microbiol 14:2457–2466
Yeomans JC, Bremner JM (1988) A rapid and precise method for routine determination of organic carbon in soil. Comm Soil Sci Plant Anal 19:1467–1476
Zhang L, Xu Z (2008) Assessing bacterial diversity in soil. J Soils Sedim 8:379–388
Zhang B, Wu X, Zhang G, Li S, Zhang W, Chen X, Sun L, Zhang B, Liu G, Chen T (2016) The diversity and biogeography of the communities of Actinobacteria in the forelands of glaciers at a continental scale. Environ Res Lett 11:054012
Acknowledgements
The authors thank “Fundação de Amparo a Pesquisa no Estado do Piauí” (FAPEPI) and “Conselho Nacional de Desenvolvimento Científico e Tecnológico” (CNPq) for financial support to this project through of PRONEX (FAPEPI/CNPq 004/2012). A.S.F. Araujo, W.M. Bezerra, A.C.A. Lopes, and V.M.M. Melo are supported by grants from CNPq (Research’s Productivity). V.M Santos is supported by grants from FAPEPI (Regional Scientific Development).
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
de Araujo, A.S.F., Bezerra, W.M., dos Santos, V.M. et al. Distinct bacterial communities across a gradient of vegetation from a preserved Brazilian Cerrado. Antonie van Leeuwenhoek 110, 457–469 (2017). https://doi.org/10.1007/s10482-016-0815-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10482-016-0815-1