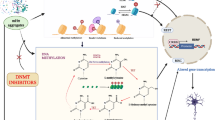
Abstract
Huntington’s disease (HD) is an incurable and fatal hereditary neurodegenerative disorder of mid-life onset characterized by chorea, emotional distress, and progressive cognitive decline. HD is caused by an expansion of CAG repeats coding for glutamine (Q) in exon 1 of the huntingtin gene. Recent studies suggest that epigenetic modifications may play a key role in HD pathogenesis. Alterations of the epigenetic “histone code” lead to chromatin remodeling and deregulation of neuronal gene transcription that are prominently linked to HD pathogenesis. Furthermore, specific noncoding RNAs and microRNAs are associated with neuronal damage in HD. In this review, we discuss how DNA methylation, post-translational modifications of histone, and noncoding RNA function are affected and involved in HD pathogenesis. In addition, we summarize the therapeutic effects of histone deacetylase inhibitors and DNA binding drugs on epigenetic modifications and neuropathological sequelae in HD. Our understanding of the role of these epigenetic mechanisms may lead to the identification of novel biological markers and new therapeutic targets to treat HD.





Similar content being viewed by others
References
Myers RH, MacDonald ME, Koroshetz WJ, et al. De novo expansion of a (CAG)n repeat in sporadic Huntington’s disease. Nat Genet 1993;5:168–173.
Kremer BP, Goldberg S, Andrew J, et al. A worldwide study of the Huntington’s disease mutation the sensitivity and specificity of measuring CAG repeats. New Engl J Med 1994;330:1401–1406.
Huntington G. On chorea. Med Surg Rep (Philadelphia) 1872;317–321.
Jergelsma G. Nue anatomische befunde bei paralysis agitans und bei chronischer progressive chorea. Neurol Centralbl 1908;27:995–996.
Bruyn GW, Bots GTAM, Dom R. Huntinton’s chorea: Current neuropathological status. In: Chase TN, Wexler NS, Barbeau A (eds) Huntington’s disease. Advances in neurology. Raven Press, New York, 1979, pp. 83–93.
Vonsattel JP, Myers RH, Stevens TJ, Ferrante RJ, Bird ED, Richardson EP Jr. Neuropathological classification of Huntington’s disease. J Neuropath Exp Neurol 1985;44:559–577.
Kowall NW, Ferrante RJ. Huntinton’s disease. In Markesbery WR (ed.) Neuropathology of dementing disorders. Oxford University Press, New York, 1998, pp. 219–256.
Hersch SM, Rosas HD, Ferrante RJ. Neuropathology and pathophysiology of Huntington’s disease in movement disorders. In: Koller W (ed.) Neurologic principles and practice. McGraw-Hill, New York, 2004, pp. 503–526.
Ferrante RJ, Kowall NW, Beal MF, Richardson EP Jr, Bird ED, Martin JB. Selective sparing of a class of striatal neurons in Huntington’s disease. Science 1985;230:561–563.
Ferrante RJ, Kowall NW, Richardson Jr. EP, Bird ED, Martin JB. Topography of encephalin, substance P and acetylcholinesterase staining in Huntington’s disease striatum. Neurosci Lett 1986;71:283–288.
Ferrante RJ, Kowall NW, Beal MF, Martin JB, Bird ED, Richardson EP Jr. Morphologic and histochemical characteristics of a spared subset of striatal neurons in Huntington’s disease. J Neuropathol Exp Neurol 1987;1:12–27.
Ferrante RJ, Kowall NW, Richardson EP, Jr. Proliferative and degenerative changes in striatal spiny neurons in Huntington’s disease: a combined study using the section-Golgi method and calbindin D28K immunocytochemistry. J Neurosci 1991;11:3877–3887.
Graveland GA, Williams RS, DiFiglia M. Evidence for degenerative and regenerative changes in neostriatal spiny neurons in Huntington’s disease. Science 1985;227:770–773.
Kowall NW, Ferrante RJ, Martin JB. Patterns of cell loss in Huntington’s disease. TINS 1987;10:24–29.
Huntington’s disease Collaborative Research Group. A novel gene containing a trinucleotide repeat that is expanded and unstable on Huntington’s disease chromosomes. Cell 1993;72:971–983.
Spada ARL, Wilson EM, Lubahn DB, Harding AE, Fischbeck KH. Androgen receptor gene mutations in X-linked spinal and bulbar muscular atrophy. Nature 1991;352:77–79.
Orr HT, Chung MY, Banfi S, et al. Expansion of an unstable trinucleotide CAG repeat in spinocerebellar ataxia type 1. Nat Genet 1993;4:221–226.
Szebenyi G, Morfini GA, Babcock A, et al. Neuropathogenic forms of huntingtin and androgen receptor inhibit fast axonal transport. Neuron 2003;40:41–52.
Trushina E, Dyer RB, Badger 2nd JD, et al. Mutant huntingtin impairs axonal trafficking in mammalian neurons in vivo and in vitro. Mol Cell Biol 2004;24:8195–8209.
Ross, CA. Intranuclear neuronal inclusions: a common pathogenic mechanism for glutamine-repeat neurodegenerative diseases? Neuron 1997;19:1147–1150.
Nucifora FC Jr, Sasaki M, Peters MF, et al. Interference by huntingtin and atrophin-1 with CBP-mediated transcription leading to cellular toxicity. Science 2001;291:2423–2428.
Cattaneo E. Dysfunction of wild-type huntingtin in Huntington disease. News Physiol Sci 2003;18:34–37.
Zhang Y, Li M, Drozda M, et al. Depletion of wild-type huntingtin in mouse models of neurologic diseases. J Neurochem 2003;87:102–106.
Ross CA, Poirier MA. Protein aggregation and neurodegenerative disease. Nat Med (Suppl.) 2004;10:S10-17.
Mangiarini L, Sathasivam K, Seller M, et al. Exon 1 of the HD gene with an expanded CAG repeat is sufficient to cause a progressive neurological phenotype in transgenic mice. Cell 87:493–506.
Yamamoto A, Lucas JJ, Hen R. Reversal of neuropathology and motor dysfunction in a corditional model of Huntington’s disease. Cell 2000;101:57–66.
Ordway JM, Tallaksen-Greene S, Gutekunst CA, et al. Ectopically expressed CAG repeats cause intranuclear inclusions and a progressive late onset neurological phenotype in the mouse. Cell 1997;91:753–763.
Beal MF, Ferrante RJ. Experimental therapeutics in transgenic mouse models of Huntington’s disease. Nat Rev Neurosci 2004;5:373–384.
Ryu H, Rosas HD, Hersch SM, Ferrante RJ. The therapeutic role of creatine in Huntington’s disease. Pharmacol Ther 2005;108:193–207.
Waddington CH. The epigenotype. Endeavour 1942;1:18.
Goldberg AD, Allis CD, Bernstein E. Epigenetics: a landscape takes shape. Cell 2007;128:635–638.
Sadri-Vakili G, Cha JH. Mechanisms of disease: Histone modifications in Huntington’s disease. Nat Clin Pract Neurol 2006;2:330–333.
Bird AP. CpG-rich islands and the function of DNA methylation. Nature 1986;321:209–213.
Ryu H, Ferrante RJ. Emerging chemotherapeutic strategies for Huntinton’s disease. Expert Opin Emerg Drugs 2005; 10: 345–363.
Boyes J, Bird A. Repression of genes by DNA methylation depends on CpG density and promoter strength: Evidence for involvement of a methyl-CpG binding protein. EMBO J 1992;11:327–333.
Lewis JD, Meehan RR, Henzel WJ, et al. Purification, sequence, and cellular localization of a novel chromosomal protein that binds to methylated DNA. Cell 1992;69:905–914.
Chan MF, van Amerongen R, Nijjar T, Cuppen E, Jones PA, Laird PW. Reduced rates of gene loss, gene silencing, and gene mutation in Dnmt1-deficient embryonic stem cells. Mol Cell Biol 2001;21:7587–7600.
Fan G, Beard C, Chen RZ, et al. DNA hypomethylation perturbs the function and survival of CNS neurons in postnatal animals. J Neurosci 2001;21:788–797.
Jackson M, Krassowska A, Gilbert N, et al. Severe global DNA hypomethylation blocks differentiation and induces histone hyperacetylation in embryonic stem cells. Mol Cell Biol 2004;24:8862–8871.
Milutinovic S, Brown SE, Zhuang Q, Szyf M. DNA methyltransferase 1 knock down induces gene expression by a mechanism independent of DNA methylation and histone deacetylation. J Biol Chem 2004;279:27915–27927.
Espada J, Ballestar E, Santoro R, et al. Epigenetic disruption of ribosomal RAN genes and nucleolar architecture in DNA methyltransferase 1 (Dnmt1) deficient cells. Nucleic Acids Res 2007;35:2191–2198.
Zhang Y, Reinberg D. Transcription regulation by histone methylation: interplay between different covalent modifications of the core histone tails. Genes Dev 2001;15:2343–2360.
Grewal SI, Jia S. Heterochromatin revisited. Nat Rev Genet 2007;8:35–46.
Ng CW, Yildirim F, Yap YS, et al. Extensive changes in DNA methylation are associated with expression of mutant huntingtin. Proc Natl Acad Sci U S A 2013;110:2354–2359.
Villar-Menéndez I, Blanch M, Tyebji S, et al. Increased 5-methylcytosine and decreased 5-hydroxymethylcytosine levels are associated with reduced striatal A(2A)R levels in Huntington’s disease. Neuromolecular Med 2013;15:295–309.
Wood H. Neurodegenerative disease: Altered DNA methylation and RNA splicing could be key mechanisms in Huntington disease. Nat Rev Neurol 2013;9:119.
Thomas B, Matson S, Chopra V, et al. A novel method for detecting 7-methyl guanine reveals aberrant methylation levels in Huntington disease. Anal Biochem 2013;436:112–120.
Hake SB, Xiao A, Allis CD. Linking the epigenetic ‘language’ of covalent histone modifications to cancer. Br J Cancer 2004;90:761–769.
Marques SC, Oliveira CR, Outeiro TF, Pereira CM. Alzheimer’s disease: the quest to understand complexity. J Alzheimers Dis 2010;21:373–383.
Chouliaras L, Rutten BP, Kenis G, et al. Epigenetic regulation in the pathophysiology of Alzheimer’s disease. Prog Neurobiol 2010;90:498–510.
Bannister AJ, Schneider R, Kouzarides T. Histone methylation: dynamic or static? Cell 2002;109:801–806.
Felsenfeld G, Groudine M. Controlling the double helix. Nature 2003;421:448–453.
Suzuki MM, Bird A. DNA methylation landscapes: provocative insights from epigenomics. Nat Rev Genet 2008;9:465–476.
Jenuwein T, Allis CD. Translating the histone code. Science 2001;293:1074–1080.
Alarcón JM, Malleret G, Touzani K, et al. Chromatin acetylation, memory, and LTP are impaired in CBP+/− mice: a model for the cognitive deficit in Rubinstein-Taybi syndrome and its amelioration. Neuron 2004;42:947–959.
Ferrante RJ, Ryu H, Kubilus JK, et al. Chemotherapy for the brain: the antitumor antibiotic mithramycin prolongs survival in a mouse model of Huntington’s disease. J Neurosci 2004;24:10335–10342.
Steffan JS, Bodai L, Pallos J, et al. Histone deacetylase inhibitors arrest polyglutamine-dependent neurodegeneration in Drosophila. Nature 2001;413:739–743.
Igarashi S, Morita H, Bennett KM, et al. Inducible PC12 cell model Huntington’s disease shows toxicity and decreased histone acetylation. Neuroreport 2003;14:565–568
Sadri-Vakili G, Bouzou B, Benn CL, et al. Histones associated with downregulated genes are hypo-acetylated in Huntington’s disease models. Hum Mol Genet 2007;16:1293–1306.
Thomas EA, Coppola G, Desplats PA, et al. The HDAC inhibitor 4b ameliorates the disease phenotype and transcriptional abnormalities in Huntington’s disease transgenic mice. Proc Natl Acad Sci U S A 2008;105:15564–15569.
McFarland KN, Das S, Sun TT, et al. Genome-wide histone acetylation is altered in a transgenic mouse model of Huntington’s disease. PLoS One 2012;7:e41423.
Bannister AJ, Kouzarides T. The CBP co-activator is a histone acetyltransferase. Nature 1996;384:641–643.
Korzus E, Rosenfeld MG, Mayford M. CBP histone acetyltransferase activity is a critical component of memory consolidation. Neuron 2004;42:961–972.
West RL, Lee JM, Maroun LE. Hypomethylation of the amyloid precursor protein gene in the brain of an Alzheimer’s disease patient. J Mol Neurosci 1995;6:141–146.
Gardian G, Browne SE, Choi DK, et al. Neuroprotective effects of phenylbutyrate in the N171-82Q transgenic mouse model of Huntington’s disease. J Biol Chem 2005;280:556–563.
Ryu H, Lee J, Hagerty SW, et al. ESET/SETDB1 gene expression and histone H3(K9) trimethylation in Huntington’s disease. Proc Natl Acad Sci U S A 2006;103:19176–19181.
Oliveira AM, Wood MA, McDonough CB, Abel T. Transgenic mice expressing an inhibitory truncated form of p300 exhibit long-term memory deficits. Learn Mem 2007;14:564–572.
Lee J, Hagerty S, Cormier KA, et al. Monoallele deletion of CBP leads to pericentromeric heterochromatin condensation through ESET expression and histone H3(K9) methylation. Hum Mol Genet 2008;17:1774–1782.
Lee J, Hwang YJ, Shin JY, et al. Epigenetic regulation of cholinergic receptor M1 (CHRM1) by histone H3K9me3 impairs Ca(2+) signaling in Huntington’s disease. Acta Neuropathol 2013;125:727–739.
Stack EC, Del Signore SJ, Luthi-Carter R, et al. Modulation of nucleosome dynamics in Huntington’s disease. Hum Mol Genet 2007;16:1164–1175.
Wu R, Terry AV, Singh PB, Gilbert DM. Differential subnuclear localization and replication timing of histone H3 lysine 9 methylation states. Mol Biol Cell 2005;16:2872–2881.
Sugita S, Uchimura N, Jiang ZG, North RA. Distinct muscarinic receptors inhibit release of GABA and excitatory amino acids in mammalian brain. Proc Natl Acad Sci U S A 1991;88:2608–2611.
Bonner TI, Buckley NJ, Young AC, Brann MR. Identification of a family of muscarinic acetylcholine receptor genes. Science 1897;237:527–532.
Hulme EC, Birdsall NJM, Buckley NJ. Muscarinic receptor subtypes. Annu Rev Pharmacol Toxicol 1990;30:633–673.
Wess J. Molecular biology of muscarinic acetylcholine receptors. Crit Rev Neurobiol 1996;10:66–69.
Calabresi P, Centonze D, Gubellini P, Pisani A, Bernardi G. Acetylcholine-mediated modulation of striatal function. Trends Neurosci 2000;23:120–126.
Cha JH, Kosinski CM, Kerner JA, et al. Altered brain neurotransmitter receptors in transgenic mice expressing a portion of an abnormal human Huntington disease gene. Proc Natl Acad Sci U S A 1998;95:6480–6485.
Wang Z, Kai L, Day M, et al. Dopaminergic control of corticostriatal long-term synaptic depression in medium spiny neurons is mediated by cholinergic interneurons. Neuron 2006;50:443–452.
Wilson CJ. Striatal D2 receptors and LTD: yes, but not where you thought they were. Neuron 2006;50:347–348.
Han J, Pedersen JS, Kwon SC, et al. Posttranscriptional crossregulation between Drosha and DGCR8. Cell 2009;136:75–84.
Han J, Lee Y, Yeom KH, Kim YK, Jin H, Kim VN. The Drosha-DGCR8 complex in primary microRNA processing. Genes Dev 2004;18:3016–3027.
Johnson R, Zuccato C, Belyaev ND, Guest DJ, Cattaneo E, Buckley NJ. A microRNA-based gene dysregulation pathway in Huntington’s disease. Neurobiol Dis 2008;29:438–445.
Lee ST, Chu K, Im WS, et al. Altered microRNA regulation in Huntington’s disease models. Exp Neurol 2011;227:172–179.
Buckley NJ, Johnson R. New insights into non-coding RNA networks in Huntington’s disease. Exp Neurol 2011;231:191–194.
Ghose J, Sinha M, Das E, Jana NR, Bhattacharyya NP. Regulation of miR-146a by RelA/NFkB and p53 in STHdhQ111/HdhQ111 cells, a cell model of Huntington’s disease. PLos One 2011;6:e23837
Bae BI, Xu H, Igarashi S, et al. p53 mediates cellular dysfunction and behavioral abnormalities in Huntington’s disease. Neuron 2005;47:29–41.
Marks PA, Richon VM, Rifkind RA. Histone deacetylase inhibitors: inducers of differentiation or apoptosis of transformed cells. J Natl Cancer Inst 2000;92:1210–1216.
Fukuda H, Sano N, Muto S, Horikoshi M. Simple histone acetylation plays a complex role in the regulation of gene expression. Brief Funct Genomic Proteomic 2006;5:190–208.
Sugai F, Yamamoto Y, Miyaguchi K, et al. Benefit of valpronic acid in suppressing disease progression of ALS model mice. Eur J Neurosci 2004;20:3179–3183.
Camelo S, Iglesias AH, Hwang D, et al. Transcriptional therapy with the histone deacetylase inhibitor trichostatin A ameliorates experimental autoimmune encephalomyelitis. J Neuroimmunol 2005;164:10–21.
Hockly E, Richon VM, Woodman B, et al. Suberoylanilide hydroxamic acid, a histone deacetylase inhibitor, ameliorates motor deficits in a mouse model of Huntington’s disease. Proc Natl Acad Sci U S A 2003;100:2041–2046.
Mielcarek M, Benn CL, Franklin SA, et al. SAHA decreases HDAC2 and 4 levels in vivo and improves molecular phenotypes in the R6/2 mouse model of Huntington’s disease. PLos One 2011;6:e27746.
McCampbell A, Taye AA, Whitty L, Penney E, Steffan JS, Fischbeck KH. Histone deacetylase inhibitors reduce polyglutamine toxicity. Proc Natl Acad Sci U S A 2001;98:15179–15184.
Ferrante RJ, Kubilus JK, Lee J, et al. Histone deacetylase inhibition by sodium butyrate chemotherapy ameliorates the neurodegenerative phenotype in Huntington’s disease mice. J Neurosci 2003;23:9418–9427.
Miller AA, Kurschel E, Osieka R, Schmidt CG. Clinical pharmacology of sodium butyrate in patients with acute leukemia. Eur J Cancer Clin Oncol 1987;23:1283–1287.
Daniel P, Brazier M, Cerutti I, et al. Pharmacokinetic study of butyric acid administered in vivo as sodium and arginine butyrate salt. Clin Chim Acta 1989;181:255–263.
Egorin MJ, Yuan ZM, Sentz DL, Plaosance K, Eiseman JL. Plasma pharmacokinetics of butyrate after intravenous administration of sodium, butyrate or oral administration of tributyrin or sodium butyrate to mice and rats. Cancer Chemother Pharmacol 1999;43:445–453.
Dasgupta S, Zhou Y, Jana M, Banik NL, Pahan K. Sodium phenylacetate inhibits adoptive transfer of experimental allergic encephaplomyelitis in SJL/J mice at multiple steps. J immunol 2003;170:3874–3882.
Hogarth P, Lovrecic L, Krainc D. Sodium phenylbutyrate in Huntington’s disease: a dose-finding study. Mov Disord 2007;22:1962–1964.
Ebbel EN, Leymarie N, Schiavo S, et al. Identification of phenylbutyrate-generated metabolites in Huntington disease patients using parallel liquid chromatography/electrochemical array/mass spectrometry and off-line tandem mass spectrometry. Anal Biochem 2010;399:152–161.
Jia H, Kast RJ, Steffan JS, Thomas EA. Selective histone deacetylase (HDAC) inhibition imparts beneficial effects in Huntington’s disease mice: implications for the ubiquitin-proteasomal and autophagy systems. Hum Mol Genet 2012;21:5280–5293.
Jia H, Pallos J, Jacques V, et al. Histone deacetylase (HDAC) inhibitors targeting HDAC3 and HDAC1 ameliorate polyglutamine-elicited phenotypes in model systems of Huntington’s disease. Neurobiol Dis 2012;46:351–361.
Hu Y, Chopra V, Chopra R, et al. Transcriptional modulator H2A histone family, member Y (H2AFY) marks Huntington disease activity in man and mouse. Proc Natl Acad Sci U S A 2011;108:17141–17146.
Blanco G, Fu H, Mendez C, Khosla C, Salas JA. Deciphering the biosynthetic origin of the aglycone of the aureolic acid group of anti-tumor agents. Chem Biol 1996;3:193–196.
Ralston SH. Pathogenesis and management of cancer associated hypercalcaemia. Cancer Surv 1994;21:179–196.
Chakrabarti S, Bhattacharyya D, Dasgupta D. Structural basis of DNA recognition by anticancer antibiotics, chromomycin A3, and mithramycin: roles of minor groove width and ligand flexibility. Biopolymers 2001;56:85–95.
Hescock H Jr, Parker M, Wang TY, Ballinger R, Balducci L. Metastatic carcinoma of unknown primary: complete response to second-line treatment with plicamycin. Am J Med Sci 1989;298:34–37.
Prado L, Lombo F, Brana AF, Mendez C, Rohr J, Salas JA. Analysis of two chromosomal region adjacent to genes for a Type II polyketide synthase involved in the biosynthesis of the antitumor polyketide mithramycin in Streptomyces argillaceus. Mol Gen Genet 1999;261:216–225.
Ryan WG. Treatment of Paget’s disease of bone with mithramycin. Clin Orthop 1977;127:106–110.
Kennedy BJ. Mithramycin therapy in testicular cancer. J Urol 1972;107:429–432.
Hagen G, Dennig J, Preiss A, Beato M, Suske G. Functional analyses of the transcription factor Sp4 reveal properties distinct from Sp1 and Sp3. J Biol Chem 1995;270:24989–24994.
Majello B, De Luca P, Suske G, Lania L. Differential transcriptional regulation of c-myc promoter through the same DNA binding sites targeted by Sp1-like proteins. Oncogene 1995;10:1841–1848.
Chatterjee S, Zaman K, Ryu H, Conforto A, Ratan RR. Sequence-selective DNA binding drugs mithramycin A and chromomycin A3 are potent inhibitors of neuronal apoptosis induced by oxidative stress and DNA damage in cortical neurons. Ann Neurol 2001;49:345–354.
Zhang Y, Friedlander RM. Using non-coding small RNAs to develop therapies for Huntington’s disease. Gene Ther 2011;18:1139–1149.
Hu J, Matsui M, Gagnon KT, et al. Allele-specific silencing of mutant huntingtin and ataxin-3 genes by targeting expanded CAG repeats in mRNAs. Nat Biotechnol 2009;27:478–484.
Lombardi MS, Jaspers L, Spronkmans C, et al. A majority of Huntington’s disease patients may be treatable by individualized allele-specific RNA interference. Exp Neurol 2009;217:312–319.
Harper SQ, Staber PD, He X, et al. RNA interference improves motor and neuropathological abnormalities in a Huntington’s disease mouse model. Proc Natl Acad Sci U S A 2005;102:5820–5825.
Franich NR, Fitzsimons HL, Fong DM, Klugmann M, During MJ, Young D. AAV vector-mediated RNAi of mutant huntingtin expression is neuroprotective in a novel genetic rat model of Huntington’s disease. Mol Ther 2008;16:947–956.
Rodriguez-Lebron E, Denovan-Wright EM, Nash K, Lewin AS, Mandel RJ. Intrastriatal rAAV-mediated delivery of anti-huntingtin shRNAs induces partial reversal of disease progression in R6/1 Huntington’s disease transgenic mice. Mol Ther 2005;12:618–633.
Machida Y, Okada T, Kurosawa M, Oyama F, Ozawa K, Nukina N. rAAV-mediated shRNA ameliorated neuropathology in Huntington disease model mouse. Biochem Biophys Res Commun 2006;343:190–197.
Huang B, Schiefer J, Sass C, Landwehrmeyer GB, Kosinski CM, Kochanek S. High capacity adenoviral vector-mediated reduction of huntingtin aggregate load in vitro and in vivo. Hum Gene Ther 2007;18:303–311.
Drouet V, Perrin V, Hassig R, et al. Sustained effects of nonallele-specific Huntingtin silencing. Ann Neurol 2009;65:276–285.
McBride JL, Boudreau RL, Harper SQ, et al. Artificial miRNAs mitigate shRNA-mediated toxicity in the brain: implications for the therapeutic development of RNAi. Proc Natl Acad Sci U S A 2008;105:5868–5873.
Boudreau RL, McBride JL, Martins I, et al. Nonallele-specific silencing of mutant and wild-type huntingtin demonstrates therapeutic efficacy in Huntington’s disease mice. Mol Ther 2009;17:1053–1063.
Difiglia M, Sena-Exteves M, Chase K, et al. Therapeutic silencing of mutant huntingtin with siRNA attenuates striatal and cortical neuropathology and behavioral deficits. Proc Natl Acad Sci U S A 2007;104:17204–17209.
Yu D, Pendergraff H, Liu J, et al. Single-stranded RNAs use RNAi to potently and allele-selectively inhibit mutant huntingtin expression. Cell 2012;150:895–908.
Luthi-Carter R, Cha J-HJ. Mechanisms of transcriptional dysregulation in Huntington’s disease. Clin Neurosci Res 2003;3:165–177.
Ryu H, Lee J, Olofsson BA, et al. Histone deacetylase inhibitors prevent oxidative neuronal death independent of expanded polyglutamine repeats via an Sp1-dependent pathway. Proc Natl Acad Sci U S A 2003;23:3597–3606.
Dompierre JP, Godin JD, Charrin BC, et al. Histone deacetylase 6 inhibition compensates for the transport deficit in Huntington’s disease by increasing tubulin acetylation. J Neurosci 2007;27:3571–3583.
Payao SL, Smith MD, Bertolucci PH. Differential chromosome sensitivity to 5-azacytidine in Alzheimer’ disease. Gerontology 1998;44:267–271.
Acknowledgments
This study was supported by NIH NS 067283–02 (H.R.), WCU Neurocytomics Program Grant (800–20080848) (H.R.), and SRC Grant (2010-0029-403) (H.R.) from National Research Foundation, and Flagship Grant (H.R.) from KIST.
Required Author Forms
Disclosure forms provided by the authors are available with the online version of this article.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
ESM 1
(PDF 499 kb)
Rights and permissions
About this article
Cite this article
Lee, J., Hwang, Y.J., Kim, K.Y. et al. Epigenetic Mechanisms of Neurodegeneration in Huntington’s Disease. Neurotherapeutics 10, 664–676 (2013). https://doi.org/10.1007/s13311-013-0206-5
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13311-013-0206-5