Abstract
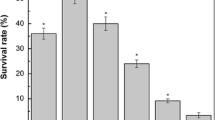
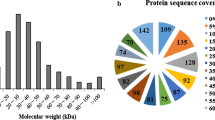
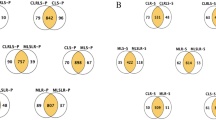
Lactobacillus casei has traditionally been recognized as a probiotic and frequently used as an adjunct culture in fermented dairy products, where acid stress is an environmental condition commonly encountered. In the present study, we carried out a comparative physiological and proteomic study to investigate lactic-acid-induced alterations in Lactobacillus casei Zhang (WT) and its acid-resistant mutant. Analysis of the physiological data showed that the mutant exhibited 33.8% higher glucose phosphoenolpyruvate:sugar phosphotransferase system activity and lower glycolytic pH compared with the WT under acidic conditions. In addition, significant differences were detected in both cells during acid stress between intracellular physiological state, including intracellular pH, H+-ATPase activity, and intracellular ATP pool. Comparison of the proteomic data based on 2D-DIGE and i-TRAQ indicated that acid stress invoked a global change in both strains. The mutant protected the cells against acid damage by regulating the expression of key proteins involved in cellular metabolism, DNA replication, RNA synthesis, translation, and some chaperones. Proteome results were validated by Lactobacillus casei displaying higher intracellular aspartate and arginine levels, and the survival at pH 3.3 was improved 1.36- and 2.10-fold by the addition of 50-mM aspartate and arginine, respectively. To our knowledge, this is the first demonstration that aspartate may be involved in acid tolerance in Lactobacillus casei. Results presented here may help us understand acid resistance mechanisms and help formulate new strategies to enhance the industrial applications of this species.









Similar content being viewed by others
References
Abdullah-Al-Mahin SS, Higashi C, Matsumoto S, Sonomoto K (2010) Improvement of multiple-stress tolerance and lactic acid production in Lactococcus lactis NZ9000 under conditions of thermal stress by heterologous expression of Escherichia coli dnaK. Appl Environ Microb 76(13):4277–4285
Belli WA, Marquis RE (1991) Adaptation of Streptococcus mutans and Enterococcus hirae to acid stress in continuous culture. Appl Environ Microb 57(4):1134–1138
Bender G, Marquis R (1987) Membrane ATPases and acid tolerance of Actinomyces viscosus and Lactobacillus casei. Appl Environ Microb 53(9):2124–2128
Breeuwer P, Drocourt J, Rombouts F, Abee T (1996) A novel method for continuous determination of the intracellular pH in bacteria with the internally conjugated fluorescent probe 5 (and 6-)-carboxyfluorescein succinimidyl ester. Appl Environ Microb 62(1):178–183
Broadbent JR, Larsen RL, Deibel V, Steele JL (2010) Physiological and transcriptional response of Lactobacillus casei ATCC 334 to acid stress. J Bacteriol 192:2445–2458
Brown ED, Vivas EI, Walsh CT, Kolter R (1995) MurA (MurZ), the enzyme that catalyzes the first committed step in peptidoglycan biosynthesis, is essential in Escherichia coli. J Bacteriol 177(14):4194–4197
Budin-Verneuil A, Pichereau V, Auffray Y, Ehrlich DS, Maguin E (2005) Proteomic characterization of the acid tolerance response in Lactococcus lactis MG1363. Proteomics 5(18):4794–4807. doi:10.1002/pmic.200401327
Carranza P, Grunau A, Schneider T, Hartmann I, Lehner A, Stephan R, Gehrig P, Grossmann J, Groebel K, Hoelzle LE, Eberl L, Riedel K (2010) A gel-free quantitative proteomics approach to investigate temperature adaptation of the food-borne pathogen Cronobacter turicensis 3032. Proteomics 10(18):3248–3261. doi:10.1002/pmic.200900460
Casiano-Colon A, Marquis R (1988) Role of the arginine deiminase system in protecting oral bacteria and an enzymatic basis for acid tolerance. Appl Environ Microb 54(6):1318–1324
Cotter PD, Hill C (2003) Surviving the acid test: responses of gram-positive bacteria to low pH. Microbiol Mol Biol R 67(3):429–453. doi:10.1128/mmbr.37.3.429-453.2003
De Angelis M, Bini L, Pallini V, Cocconcelli PS, Gobbetti M (2001) The acid-stress response in Lactobacillus sanfranciscensis CB1. Microbiol 147:1863–1873
De Angelis M, Di Cagno R, Huet C, Crecchio C, Fox P, Gobbetti M (2004) Heat shock response in Lactobacillus plantarum. Appl Environ Microb 70(3):1336–1346
De Angelis M, Gobbetti M (2004) Environmental stress responses in Lactobacillus: a review. Proteomics 4(1):106–122
Desmond C, Fitzgerald GF, Stanton C, Ross RP (2004) Improved stress tolerance of GroESL-overproducing Lactococcus lactis and probiotic Lactobacillus paracasei NFBC 338. Appl Environ Microb 70(10):5929–5936. doi:10.1128/aem.70.10.5929-5936.2004
Fernández M, Zú iga M (2006) Amino acid catabolic pathways of lactic acid bacteria. Crit Rev Microbiol 32(3):155–183
Fountoulakis M, Lahm H (1998) Hydrolysis and amino acid composition analysis of proteins. J Chromatogr A 826(2):109–134
Friedberg E (2003) DNA damage and repair. Nature 421(6921):436–440
Greenacre EJ, Brocklehurst TF (2006) The acetic acid tolerance response induces cross-protection to salt stress in Salmonella typhimurium. Int J Food Microbiol 112(1):62–65. doi:10.1016/j.ijfoodmicro.2006.05.012
Hartke A, Bouche S, Giard JC, Benachour A, Boutibonnes P, Auffray Y (1996) The lactic acid stress response of Lactococcus lactis subsp. lactis. Curr Microbiol 33(3):194–199
Hormann S, Scheyhing C, Behr J, Pavlovic M, Ehrmann M, Vogel R (2006) Comparative proteome approach to characterize the high-pressure stress response of Lactobacillus sanfranciscensis DSM 20451T. Proteomics 6(6):1878–1885
Hutkins RW, Nannen NL (1993) pH homeostasis in lactic acid bacteria. J Dairy Sci 76(8):2354–2365
Jung YJ, Min KJ, Yoon KS (2009) Responses of acid-stressed Salmonella typhimurium in broth and chicken patties to subsequent antimicrobial stress with ε-polylysine and combined potassium lactate and sodium diacetate. Food Microbiol 26:467–474
Kleerebezem M, Vaughan EE (2009) Probiotic and gut lactobacilli and bifidobacteria: molecular approaches to study diversity and activity. Annu Rev Microbiol 63:269–290. doi:10.1146/annurev.micro.091208.073341
Kornberg H, Reeves R (1972) Inducible phosphoenolpyruvate-dependent hexose phosphotransferase activities in Escherichia coli. Biochem J 128(5):1339–1344
Lambert R, Stratford M (1999) Weak-acid preservatives: modelling microbial inhibition and response. J Appl Microbiol 86(1):157–164
Langereis J, Prinsen B, Coppens C, Koenderman L, Ulfman L (2009) A 2D-DIGE approach to identify proteins involved in inside-out control of integrins. J Proteome Res 8(11):3824–3833
Lebeer S, Vanderleyden J, De Keersmaecker SCJ (2008) Genes and molecules of lactobacilli supporting probiotic action. Microbiol Mol Biol R 72(4):728–764
LeBlanc D, Crow V, Lee L, Garon C (1979) Influence of the lactose plasmid on the metabolism of galactose by Streptococcus lactis. J Bacteriol 137(2):878–884
Lee KB, Lee HG, Pi KB, Choi YJ (2008) The effect of low pH on protein expression by the probiotic bacterium Lactobacillus reuteri. Proteomics 8(8):1624–1630
Len ACL, Harty DWS, Jacques NA (2004a) Stress-responsive proteins are upregulated in Streptococcus mutans during acid tolerance. Microbiol 150:1339–1351. doi:10.1099/mic.0.27008-0
Len MCL, Harty DWS, Jacques NA (2004b) Proteome analysis of Streptococcus mutans metabolic phenotype during acid tolerance. Microbiol 150:1353–1366. doi:10.1099/mic.0.26888-0
Leverrier P, Vissers J, Rouault A, Boyaval P, Jan G (2004) Mass spectrometry proteomic analysis of stress adaptation reveals both common and distinct response pathways in Propionibacterium freudenreichii. Arch Microbiol 181(3):215–230
MacLellan SR, Helmann JD, Antelmann H (2009) The YvrI alternative sigma factor is essential for acid stress induction of oxalate decarboxylase in Bacillus subtilis. J Bacteriol 191(3):931–939. doi:10.1128/jb.01435-08
Matsui R, Cvitkovitch D (2010) Acid tolerance mechanisms utilized by Streptococcus mutans. Future Microbiol 5(3):403–417
Maurer L, Yohannes E, Bondurant S, Radmacher M, Slonczewski J (2005) pH regulates genes for flagellar motility, catabolism, and oxidative stress in Escherichia coli K-12. J Bacteriol 187(1):304–319
Men H, Park P, Ge M, Walker S (1998) Substrate synthesis and activity assay for MurG. J Am Chem Soc 120(10):2484–2485
Nascimento MM, Lemos JAC, Abranches J, Goncalves RB, Burne RA (2004) Adaptive acid tolerance response of Streptococcus sobrinus. J Bacteriol 186(19):6383–6390
O’Driscoll B, Gahan C, Hill C (1997) Two-dimensional polyacrylamide gel electrophoresis analysis of the acid tolerance response in Listeria monocytogenes LO28. Appl Environ Microb 63(7):2679–2685
O’Sullivan E, Condon S (1997) Intracellular pH is a major factor in the induction of tolerance to acid and other stresses in Lactococcus lactis. Appl Environ Microb 63(11):4210–4215
Odriscoll B, Gahan CGM, Hill C (1996) Adaptive acid tolerance response in Listeria monocytogenes: isolation of an acid-tolerant mutant which demonstrates increased virulence. Appl Environ Microb 62(5):1693–1698
Parvez S, Malik KA, Kang SA, Kim HY (2006) Probiotics and their fermented food products are beneficial for health. J Appl Microbiol 100(6):1171–1185. doi:10.1111/j.1365-2672.2006.02963.x
Peran L, Camuesco D, Comalada M, Bailon E, Henriksson A, Xaus J, Zarzuelo A, Galvez J (2007) A comparative study of the preventative effects exerted by three probiotics, Bifidobacterium lactis, Lactobacillus casei and Lactobacillus acidophilus, in the TNBS model of rat colitis. J Appl Microbiol 103(4):836–844
Postma P, Lengeler J, Jacobson G (1993) Phosphoenolpyruvate: carbohydrate phosphotransferase systems of bacteria. Microbiol Mol Biol R 57(3):543–594
Randazzo CL, Restuccia C, Romano AD, Caggia C (2004) Lactobacillus casei, dominant species in naturally fermented Sicilian green olives. Int J Food Microbiol 90(1):9–14
Senouci-Rezkallah K, Schmitt P, Jobin M (2011) Amino acids improve acid tolerance and internal pH maintenance in Bacillus cereus ATCC14579 strain. Food Microbiol 28:364–372
Serrazanetti DI, Guerzoni ME, Corsetti A, Vogel R (2009) Metabolic impact and potential exploitation of the stress reactions in lactobacilli. Food Microbiol 26(7):700–711
Siegumfeldt H, Rechinger KB, Jakobsen M (2000) Dynamic changes of intracellular pH in individual lactic acid bacterium cells in response to a rapid drop in extracellular pH. Appl Environ Microb 66(6):2330–2335
Sixma T (2001) DNA mismatch repair: MutS structures bound to mismatches. Curr Opin Struc Biol 11(1):47–52
Spoering A, Vulic M, Lewis K (2006) GlpD and PlsB participate in persister cell formation in Escherichia coli. J Bacteriol 188(14):5136–5144
Streit F, Delettre J, Corrieu G, Beal C (2008) Acid adaptation of Lactobacillus delbrueckii subsp. bulgaricus induces physiological responses at membrane and cytosolic levels that improves cryotolerance. J Appl Microbiol 105 (4):1071–1080. doi:10.1111/j.1365-2672.2008.03848.x
Takahashi N, Xiao JZ, Miyaji K, Yaeshiima T, Hiramatsu A, Iwatsuki K, Kokubo S, Hosono A (2004) Selection of acid tolerant bifidobacteria and evidence for a low-pH-inducible acid tolerance response in Bifidobacterium longum. J Dairy Res 71(3):340–345. doi:10.1017/s0022029904000251
Vadeboncoeur C, Pelletier M (1997) The phosphoenolpyruvate: sugar phosphotransferase system of oral streptococci and its role in the control of sugar metabolism. FEMS Microbiol Rev 19(3):187–207
Vadeboncoeur C, St Martin S, Brochu D, Hamilton I (1991) Effect of growth rate and pH on intracellular levels and activities of the components of the phosphoenolpyruvate: sugar phosphotransferase system in Streptococcus mutans Ingbritt. Infect Immun 59(3):900–906
Viana R, Pérez-Martínez G, Deutscher J, Monedero V (2005) The glycolytic genes pfk and pyk from Lactobacillus casei are induced by sugars transported by the phosphoenolpyruvate: sugar phosphotransferase system and repressed by CcpA. Arch Microbiol 183(6):385–393
Vrancken G, Rimaux T, Weckx S, De Vuyst L, Leroy F (2009) Environmental pH determines citrulline and ornithine release through the arginine deiminase pathway in Lactobacillus fermentum IMDO 130101. Int J Food Microbiol 135(3):216–222
Waddington L, Cyr T, Hefford M, Hansen L, Kalmokoff M (2010) Understanding the acid tolerance response of bifidobacteria. J Appl Microbiol 108(4):1408–1420
Warnecke T, Gill RT (2005) Organic acid toxicity, tolerance, and production in Escherichia coli biorefining applications. Microb Cell Fact 4(1):25–32
Wilkins JC, Beighton D, Homer KA (2003) Effect of acidic pH on expression of surface-associated proteins of Streptococcus oralis. Appl Environ Microb 69(9):5290–5296
Wu R, Wang W, Yu D, Zhang W, Li Y, Sun Z, Wu J, Meng H, Zhang H (2009) Proteomic analysis of Lactobacillus casei Zhang, a new probiotic bacterium isolated from traditionally home-made Koumiss in Inner Mongolia of China. Mol Cell Proteomics 10:2321–2338
Zhang YM, Rock CO (2008) Membrane lipid homeostasis in bacteria. Nat Rev Microbiol 6(3):222–233
Zhu Y, Zhang Y, Li Y (2009) Understanding the industrial application potential of lactic acid bacteria through genomics. Appl Microbiol Biot 83(4):597–610
Acknowledgements
This project was financially supported by the Key Program of the National Natural Science Foundation of China (No. 20836003), the National Natural Science Foundation of China (No. 30900013), Major State Basic Research Development Program of China (973 Program, No. 2010CB535014), and the Self-determined Research Program of Jiangnan University(JUSRP 21009).
Author information
Authors and Affiliations
Corresponding authors
Additional information
Chongde Wu and Juan Zhang contributed equally to this paper.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Fig. S1
2D-gels for spot-picker. A total of 1200 μμg of protein was focused on 24 cm pH 4–7 IEF strips and stained with Coomassie blue R-250. Indicated protein spots were picked and subjected to in-gel digestion and mass spectrometry. (DOC 426 kb)
Fig. S2
Hierarchical clustering of proteomic data arranged by functional categories according to the COG classification. C, Energy production and conversion; D, Cell-cycle control, mitosis and meiosis; E, Amino acid transporter and metabolism; F, Nucleotide transport and metabolism; G, Carbohydrate transport and metabolism; H, Coenzyme transport and metabolism; I, Lipid transport and metabolism; J, Translation, ribosomal structure and biogenesis; K, Transcription; L, Replication recombination and repair; M, Cell wall/membrane biogenesis; O, Posttranslational modification, protein turnover, chaperones; P, Inorganic ion transport and metabolism; R, General function prediction only; UN, Function unknown and not in COG. (DOC 311 kb)
Rights and permissions
About this article
Cite this article
Wu, C., Zhang, J., Chen, W. et al. A combined physiological and proteomic approach to reveal lactic-acid-induced alterations in Lactobacillus casei Zhang and its mutant with enhanced lactic acid tolerance. Appl Microbiol Biotechnol 93, 707–722 (2012). https://doi.org/10.1007/s00253-011-3757-6
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-011-3757-6