Abstract
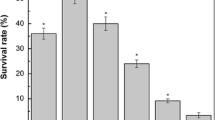
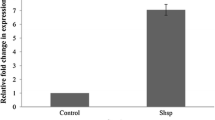
Microorganisms used in food technology and probiotics are exposed to technological and digestive stresses, respectively. Traditionally used as Swiss-type cheese starters, propionibacteria also constitute promising human probiotics. Stress tolerance and cross-protection in Propionibacterium freudenreichii were thus examined after exposure to heat, acid, or bile salts stresses. Adapted cells demonstrated acquired homologous tolerance. Cross-protection between bile salts and heat adaptation was demonstrated. By contrast, bile salts pretreatment sensitized cells to acid challenge and vice versa. Surprisingly, heat and acid responses did not present significant cross-protection in P. freudenreichii. During adaptations, important changes in cellular protein synthesis were observed using two-dimensional electrophoresis. While global protein synthesis decreased, several proteins were overexpressed during stress adaptations. Thirty-four proteins were induced by acid pretreatment, 34 by bile salts pretreatment, and 26 by heat pretreatment. Six proteins are common to all stresses and represent general stress-response components. Among these polypeptides, general stress chaperones, and proteins involved in energetic metabolism, oxidative stress response, or SOS response were identified. These results bring new insight into the tolerance of P. freudenreichii to heat, acid, and bile salts, and should be taken into consideration in the development of probiotic preparations.



Similar content being viewed by others
References
Antelmann H, Bernhardt J, Schmid R, Mach H, Volker U, Hecker M (1997) First steps from a two-dimensional protein index towards a response-regulation map for Bacillus subtilis. Electrophoresis 18:1451–1463
Bentley SD, Chater KF, Cerdeno-Tarraga AM, Challis GL, Thomson NR, James KD, Harris DE, Quail MA, Kieser H, Harper D, Bateman A, Brown S, Chandra G, Chen CW, Collins M, Cronin A, Fraser A, Goble A, Hidalgo J, Hornsby T, Howarth S, Huang CH, Kieser T, Larke L, Murphy L, Oliver K, O’Neil S, Rabbinowitsch E, Rajandream MA, Rutherford K, Rutter S, Seeger K, Saunders D, Sharp S, Squares R, Squares S, Taylor K, Warren T, Wietzorrek A, Woodward J, Barrell BG, Parkhill J, Hopwood DA (2002) Complete genome sequence of the model actinomycete Streptomyces coelicolor A3(2). Nature 417:141–147
Bernstein C, Bernstein H, Payne CM, Beard SE, Schneider J (1999) Bile salt activation of stress response promoters in Escherichia coli. Curr Microbiol 39:68–72
Bougle D, Roland N, Lebeurrier F, Arhan P (1999) Effect of propionibacteria supplementation on fecal bifidobacteria and segmental colonic transit time in healthy human subjects. Scand J Gastroenterol 34:144–148
Breton YL, Maze A, Hartke A, Lemarinier S, Auffray Y, Rince A (2002) Isolation and characterization of bile salts-sensitive mutants of Enterococcus faecalis. Curr Microbiol 45:434–439
Caldas TD, El Yaagoubi A, Richarme G (1998) Chaperone properties of bacterial elongation factor EF-Tu. J Biol Chem 273:11478–11482
Chou LS, Weimer B (1999) Isolation and characterization of acid- and bile-tolerant isolates from strains of Lactobacillus acidophilus. J Dairy Sci 82:23–31
Davis MJ, Coote PJ, O’Byrne CP (1996) Acid tolerance in Listeria monocytogenes: the adaptive acid tolerance response (ATR) and growth-phase-dependent acid resistance. Microbiology 142:2975–2982
Duche O, Tremoulet F, Glaser P, Labadie J (2002) Salt Stress Proteins Induced in Listeria monocytogenes. Appl Environ Microbiol 68:1491–1498
Duwat P (1999) Stress response pathways in Lactococcus lactis. Recent Res Devel Microbiology 3:335–348
Flahaut S, Frere J, Boutibonnes P, Auffray Y (1996a) Comparison of the bile salts and sodium dodecyl sulfate stress responses in Enterococcus faecalis. Appl Environ Microbiol 62:2416–2420
Flahaut S, Hartke A, Giard JC, Benachour A, Boutibonnes P, Auffray Y (1996b) Relationship between stress response toward bile salts, acid and heat treatment in Enterococcus faecalis. FEMS Microbiol Lett 138:49–54
Flahaut S, Frere J, Boutibonnes P, Auffray Y (1997) Relationship between the thermotolerance and the increase of DnaK and GroEL synthesis in Enterococcus faecalis ATCC19433. J Basic Microbiol 37:251–258
Foster JW (1993) The acid tolerance response of Salmonella typhimurium involves transient synthesis of key acid shock proteins. J Bacteriol 175:1981–1987
Franklyn KM, Warmington JR (1994) The expression of Candida albicans enolase is not heat shock inducible. FEMS Microbiol Lett 118:219–225
Fuller R (1989) Probiotics in man and animals. J Appl Bacteriol 66:365–378
Goldin BR and Gorbach SL (1992) Probiotics for humans. In: Fuller, R. (ed) Probiotics, the scientific basis. Chapman & Hall, London, pp 355–376
Gouesbet G, Jan G, Boyaval P (2002) Two-dimensional electrophoresis study of Lactobacillus delbrueckii subsp. bulgaricus thermotolerance. Appl Environ Microbiol 68:1055–1063
Graumann P, Schroder K, Schmid R, Marahiel MA (1996) Cold shock stress-induced proteins in Bacillus subtilis. J Bacteriol 178:4611–4619
Hartke A, Bouché S, Giard JC, Benachour A, Boutibonnes P, Auffray Y (1996) The lactic acid stress response of Lactococcus lactis subsp. lactis. Curr Microbiol 33:194–199
Hecker M, Schumann W, Volker U (1996) Heat-shock and general stress response in Bacillus subtilis. Mol Microbiol 19:417–428
Henzel WJ, Billeci TM, Stults JT, Wong SC, Grimley C, Watanabe C (1993) Identifying proteins from two-dimensional gels by molecular mass searching of peptide fragments in protein sequence databases. Proc Natl Acad Sci U S A 90:5011–5015
Hermann T, Pfefferle W, Baumann C, Busker E, Schaffer S, Bott M, Sahm H, Dusch N, Kalinowski J, Puhler A, Bendt AK, Kramer R, Burkovski A (2001) Proteome analysis of Corynebacterium glutamicum. Electrophoresis 22:1712–1723
Ikeda H, Ishikawa J, Hanamoto A, Shinose M, Kikuchi H, Shiba T, Sakaki Y, Hattori M, Omura S (2003) Complete genome sequence and comparative analysis of the industrial microorganism Streptomyces avermitilis. Nat Biotechnol 21:526–531
Jacobsen CN, Rosenfeldt N, V, Hayford AE, Moller PL, Michaelsen KF, Paerregaard A, Sandstrom B, Tvede M, Jakobsen M (1999) Screening of probiotic activities of forty-seven strains of Lactobacillus spp. by in vitro techniques and evaluation of the colonization ability of five selected strains in humans. Appl Environ Microbiol 65:4949–4956
Jan G, Leverrier P, Roland N (2001a) Survival and beneficial effects of propionibacteria in the human gut: in vivo and in vitro investigations. Lait 82:131–144
Jan G, Leverrier P, Pichereau V, Boyaval P (2001b) Changes in protein synthesis and morphology during acid adaptation of Propionibacterium freudenreichii. Appl Environ Microbiol 67:2029–2036
Jan G, Belzacq AS, Haouzi D, Rouault A, Metivier D, Kroemer G, Brenner C (2002) Propionibacteria induce apoptosis of colorectal carcinoma cells via short-chain fatty acids acting on mitochondria. Cell Death Differ 9:179–188
Jungblut PR, Bumann D, Haas G, Zimny-Arndt U, Holland P, Lamer S, Siejak F, Aebischer A, Meyer TF (2000) Comparative proteome analysis of Helicobacter pylori. Mol Microbiol 36:710–725
Laport MS, de Castro AC, Villardo A, Lemos JA, Bastos MC, Giambiagi-deMarval M (2001) Expression of the major heat shock proteins DnaK and GroEL in Streptococcus pyogenes: a comparison to Enterococcus faecalis and Staphylococcus aureus. Curr Microbiol 42:264–268
Leverrier P, Dimova D, Pichereau V, Auffray Y, Boyaval P, Jan G (2003) Susceptibility and adaptive response to bile salts in Propionibacterium freudenreichii: physiological and proteomic analysis. Appl Environ Microbiol 69:3809–3818
Lorca GL, Raya RR, Taranto MP, De Valdez GF (1998) Adaptative acid tolerance response in Lactobacillus acidophilus. Biotechnol Lett 20:239–241
Lou Y, Yousef AE (1997) Adaptation to sublethal environmental stresses protects Listeria monocytogenes against lethal preservation factors. Appl Environ Microbiol 63:1252–1255
Lyon WJ, Glatz BA (1993) Isolation and purification of propionicin PLG-1, a bacteriocin produced by a strain of Propionibacterium thoenii. Appl Environ Microbiol 59:83–88
Ma D, Cook DN, Alberti M, Pon NG, Nikaido H, Hearst JE (1995) Genes acrA and acrB encode a stress-induced efflux system of Escherichia coli. Mol Microbiol 16:45–55
Mackey AJ, Haystead TA, Pearson WR (2002) Getting more from less: algorithms for rapid protein identification with multiple short peptide sequences. Mol Cell Proteomics 1:139–147
Malik AC, Reinbold GW, Vedamuthu ER (1968) An evaluation of the taxonomy of Propionibacterium. Can J Microbiol 14:1185–1191
Mantere-Alhonen S (1995) Propionibacteria used as probiotics—A review. Lait 75:447–452
Mooney C, Munster DJ, Bagshaw PF, Allardyce RA (1990) Helicobacter pylori acid resistance. Lancet 335:1232
Mori H, Sato Y, Taketomo N, Kamiyama T, Yoshiyama Y, Meguro S, Sato H, Kaneko T (1997) Isolation and structural identification of bifidogenic growth stimulator produced by Propionibacterium freudenreichii . J Dairy Sci 80:1959–1964
Moulis JM, Davasse V, Meyer J, Gaillard J (1996) Molecular mechanism of pyruvate-ferredoxin oxidoreductases based on data obtained with the Clostridium pasteurianum enzyme. FEBS Lett 380:287–290
Murtif VL, Bahler CR, Samols D (1985) Cloning and expression of the 1.3S biotin-containing subunit of transcarboxylase. Proc Natl Acad Sci U S A 82:5617–5621
O’Sullivan E, Condon S (1997) Intracellular pH is a major factor in the induction of tolerance to acid and other stresses in Lactococcus lactis. Appl Environ Microbiol 63:4210–4215
Pérez Chaia A, Zarate G, Oliver G (1999) The probiotic properties of propionibacteria. Lait 79:175–185
Periago PM, van Schaik W, Abee T, Wouters JA (2002) Identification of proteins involved in the heat stress response of Bacillus cereus ATCC 14579. Appl Environ Microbiol 68:3486–3495
Perrot F, Hebraud M, Charlionet R, Junter GA, Jouenne T (2001) Cell immobilization induces changes in the protein response of Escherichia coli K-12 to a cold shock. Electrophoresis 22:2110–2119
Petersohn A, Brigulla M, Haas S, Hoheisel JD, Volker U, Hecker M (2001) Global analysis of the general stress response of Bacillus subtilis. J Bacteriol 183:5617–5631
Segal G, Ron EZ (1998) Regulation of heat-shock response in bacteria. Ann NY Acad Sci 851:147–151
Shah NP (2000) Probiotic bacteria: selective enumeration and survival in dairy foods. J Dairy Sci 83:894–907
Thanassi DG, Cheng LW, Nikaido H (1997) Active efflux of bile salts by Escherichia coli. J Bacteriol 179:2512–2518
Thierry A, Salvat-Brunaud D, Madec MN, Michel F, Maubois JL (1998) Affinage de l’emmental: dynamique des populations bactériennes et évolution de la composition de la phase aqueuse. Lait 78:521–542
Wilkins JC, Homer KA, Beighton D (2002) Analysis of Streptococcus mutans proteins modulated by culture under acidic conditions. Appl Environ Microbiol 68:2382–2390
Yura T, Nagai H, Mori H (1993) Regulation of the heat-shock response in bacteria. Annu Rev Microbiol 47:321–350
Zarate G, Chaia AP, Gonzalez S, Oliver G (2000) Viability and beta-galactosidase activity of dairy propionibacteria subjected to digestion by artificial gastric and intestinal fluids. J Food Prot 63:1214–1221
Acknowledgements
Standa Industrie is greatly acknowledged for financial support and for constant interest and enthusiasm in this work. P. Leverrier is the recipient of a grant from the Institut National de la Recherche Agronomique and from Standa Industrie. John Hannon is acknowledged for English correction.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Leverrier, P., Vissers, J.P.C., Rouault, A. et al. Mass spectrometry proteomic analysis of stress adaptation reveals both common and distinct response pathways in Propionibacterium freudenreichii . Arch Microbiol 181, 215–230 (2004). https://doi.org/10.1007/s00203-003-0646-0
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00203-003-0646-0