Abstract
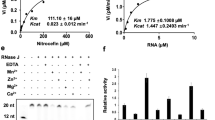
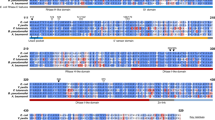
Rapid adaptations to various environmental changes are key to the survival of bacterial cells, which is mediated at the level of transcription initiation and RNA degradation/processing. Ribonuclease E (RNase E), an essential endoribonuclease gene in Mycobacterium species, is a key mediator in most reactions of RNA metabolism such as messenger RNA (mRNA), ribosomal RNA (rRNA) and transfer RNA (tRNA) processing and their degradation qualifying it as an important drug target. Here, we first report the cloning of full-length Mycobacterium smegmatis mc2155 (M. smegmatis) RNase E [MSMEG_4626] and its expression with isopropyl β-D-1-thiogalactopyranoside (IPTG) in Escherichia coli (E. coli) as a glutathione S-transferase (GST) fusion protein. The best expression of the recombinant protein was optimized with respect to different parameters, such as E. coli strains, temperature, IPTG concentrations and time points of induction. Despite the use of protease inhibitors, we observed low-molecular-weight degradation products along with full-length fusion proteins. Maximum expression of soluble and least degraded full-length RNase E was obtained at 32 °C/0.1-mM IPTG, further purified by affinity chromatography using glutathione agarose beads and confirmed by immunoblotting technique using anti-GST antibody. In silico characterization using multiple sequence alignment, homology modeling and docking studies revealed four amino acid residues, Asp613, Phe657, Ile662 and Glu688, in the catalytic domain of RNase E that are completely conserved in both E. coli and Mycobacterium species, also involved in RNA binding and might act as candidate drug targets to combat antibiotic-resistant mycobacterial diseases. Additionally, recombinant GST-RNase E protein can unravel the mechanism of RNA degradation/processing through protein–protein interaction studies.





Similar content being viewed by others
Data Availability
Not applicable.
References
Agarwala R, Barrett T, Beck J et al (2018) Database resources of the national centre for biotechnology information. Nucleic Acids Res 46(D1):D8–D13. https://doi.org/10.1093/nar/gkx1095
Alipour M, Gargari SLM, Rasooli I (2009) Cloning, expression and immunogenicity of ferric enterobactin binding protein Fep B from Escherichia coli O157: H7. Indian J Microbiol 49(3):266–270. https://doi.org/10.1007/s12088-009-0044-7
Amann E, Brosius J, Ptashne M (1983) Vectors bearing a hybrid trp-lac promoter useful for regulated expression of cloned genes in Escherichia coli. Gene 25:167–178. https://doi.org/10.1016/0378-1119(83)90222-6
Arraiano CM, Andrade JM, Domingues S, Guinote IB, Malecki M, Matos RG, Viegas SC (2010) The critical role of RNA processing and degradation in the control of gene expression. FEMS Microbiol Rev 34(5):883–923. https://doi.org/10.1111/j.1574-6976.2010.00242
Belisle JT, Sonnenberg MG (1998) Isolation of genomic DNA from mycobacteria. Methods Mol Biol, In Mycobacteria protocols. https://doi.org/10.1385/0-89603-471-2:31
Benito A, Ribó M, Vilanova M (2005) On the track of antitumour ribonucleases. MolBiosyst 1(4):294–302. https://doi.org/10.1039/b502847g
Brunelle JL, Green R (2014) One-dimensional SDS-polyacrylamide gel electrophoresis (1D SDS-PAGE). Meth Enzymol 541:151–159. https://doi.org/10.1016/B978-0-12-420119-4
Cafaro V, De Lorenzo C, Piccoli R, Bracale A, Mastronicola MR, Di Donato A, D’Alessio G (1995) The antitumor action of seminal ribonuclease and its quaternary conformations. FEBS Lett 359(1):31–34. https://doi.org/10.1016/0014-5793(94)01450-f
Callaghan AJ, Marcaida MJ, Stead JA, McDowall KJ, Scott WG, Luisi BF (2005) Structure of Escherichia coli RNase E catalytic domain and implications for RNA turnover. Nat Commun 437(7062):1187–1191. https://doi.org/10.1038/nature04084
Carpousis AJ, Luisi BF, McDowall KJ (2009) Endonucleolytic initiation of mRNA decay in Escherichia coli. Prog Mol Biol Transl Sci 85:91–135. https://doi.org/10.1016/S0079-6603(08)00803-9
Condon C, Squires C, Squires CL (1995) Control of rRNA transcription in Escherichia coli. Microbiol Rev 59(4):623–645. https://doi.org/10.1128/mr.59.4.623-645
Csanadi A, Faludi I, Miczak A (2009) MSMEG_4626 ribonuclease from Mycobacterium smegmatis. Mol Biol Rep 36(8):2341. https://doi.org/10.1007/s11033-009-9454-1
De Boer HA, Comstock LJ, Vasser M (1983) The tac promoter: a functional hybrid derived from the trp and lac promoters. Proc Natl Acad Sci 80(1):21–25. https://doi.org/10.1073/pnas.80.1.21
Deutscher MP (2015) How bacterial cells keep ribonucleases under control. FEMS Microbiol Rev 39(3):350–361. https://doi.org/10.1093/femsre/fuv012
Dumon-Seignovert L, Cariot G, Vuillard L (2004) The toxicity of recombinant proteins in Escherichia coli: a comparison of overexpression in BL21(DE3), C41(DE3), and C43(DE3). Protein Expr Purif 37(1):203–206. https://doi.org/10.1016/j.pep.2004.04.025
Gopal GJ, Kumar A (2013) Strategies for the production of recombinant protein in Escherichia coli. The Protein J 32(6):419–425. https://doi.org/10.1007/s10930-013-9502-5
Gupta A, Rath PC (2012) Expression, purification and characterization of the interferon-inducible, antiviral and tumour-suppressor protein, human RNase L. J Biosci 37(1):103–113. https://doi.org/10.1007/s12038-011-9180-4
Gupta A, Rath PC (2014b) Expression of mRNA and protein–protein interaction of the antiviral endoribonuclease RNase L in mouse spleen. Int J Biol Macromol 69:307–318. https://doi.org/10.1016/j.ijbiomac.2014.04.042
Gupta N, Gupta A, Kumar S, Mishra R, Singh C, Tripathi AK (2014a) Cross-talk between cognate and noncognate RpoE sigma factors and Zn2+-binding anti-sigma factors regulates photooxidative stress response in Azospirillumbrasilense. Antioxid Redox Signal 20(1):42–59. https://doi.org/10.1089/ars.2013.5314
Gupta RS, Lo B, Son J (2018) Phylogenomics and comparative genomic studies robustly support division of the genus Mycobacterium into an emended genus Mycobacterium and four novel genera. Front Microbiol 9:67. https://doi.org/10.3389/fmicb.2018.00067
Hadjeras L, Poljak L, Bouvier M, Morin-Ogier Q, Canal I, Cocaign-Bousquet M, Carpousis AJ (2019) Detachment of the RNA degradosome from the inner membrane of Escherichia coli results in a global slowdown of mRNA degradation, proteolysis of RNase E and increased turnover of ribosome-free transcripts. Mol Microbiol 111(6):1731. https://doi.org/10.1111/mmi.14248
Hu X, Shi Q, Yang T, Jackowski G (1996) Specific replacement of consecutive AGG codons results in high-level expression of human cardiac troponin T in Escherichia coli. Protein Expr Purif 7(3):289–293. https://doi.org/10.1006/prep.1996.0041
Joseph BC, Pichaimuthu S, Srimeenakshi S, Murthy M, Selvakumar K, Ganesan M, Manjunath SR (2015) An overview of the parameters for recombinant protein expression in Escherichia coli. J Cell SciTher 6(5):221. https://doi.org/10.4172/2157-7013.1000221
Kelley LA, Mezulis S, Yates CM, Wass MN, Sternberg MJE (2015) The Phyre2 web portal for protein modelling, prediction and analysis. Nat Protoc 10(6):845–858. https://doi.org/10.1038/nprot.2015.053
Kime L, Vincent HA, Gendoo D, Jourdan SS, Fishwick CW, Callaghan AJ, McDowall KJ (2015) The first small-molecule inhibitors of members of the ribonuclease E family. Sci Rep 5(1):1–8. https://doi.org/10.1038/srep08028
Koslover DJ, Callaghan AJ, Marcaida MJ, Garman EF, Martick M, Scott WG, Luisi BF (2008) The crystal structure of the Escherichia coli RNase E apoprotein and a mechanism for RNA degradation. Structure 16(8):1238–1244. https://doi.org/10.1016/j.str.2008.04.017
Kovacs L, Csanadi A, Megyeri K, Kaberdin VR, Miczak A (2005) Mycobacterial RNase E-associated proteins. Microbiol Immunol 49(11):1003–1007. https://doi.org/10.1111/j.1348-0421.2005.tb03697
Mackie GA (2013) RNase E: at the interface of bacterial RNA processing and decay. Nat Rev Microbiol 11(1):45–57. https://doi.org/10.1038/nrmicro2930
Mandal M, Breaker RR (2004) Gene regulation by riboswitches. Nat Rev Mol Cell Biol 5(6):451–463. https://doi.org/10.1038/nrm1403
Mardle CE, Goddard LR, Spelman BC, Atkins HS, Butt LE, Cox PA, Callaghan AJ (2020) Identification and analysis of novel small molecule inhibitors of RNase E: implications for antibacterial targeting and regulation of RNase E. Biochem Biophys Rep 23:100773. https://doi.org/10.1016/j.bbrep.2020.100773
Mukherjee R, Gomez M, Jayaraman N, Smith I, Chatterji D (2005) Hyperglycosylation of glycopeptidolipid of Mycobacterium smegmatis under nutrient starvation: structural studies. Microbiology 151(7):2385–2392. https://doi.org/10.1099/mic.0.27908-0
Płociński P, Macios M, Houghton J, Niemiec E, Płocińska R, Brzostek A, Dziembowski A (2019) Proteomic and transcriptomic experiments reveal an essential role of RNA degradosome complexes in shaping the transcriptome of Mycobacterium tuberculosis. Nucleic Acids Res 47(11):5892–5905. https://doi.org/10.1093/nar/gkz251
Rosano GL, Ceccarelli EA (2014) Recombinant protein expression in Escherichia coli: advances and challenges. Front Microbiol 5:172. https://doi.org/10.3389/fmicb.2014.00172.eCollection2014
Santosh KM, Nitish K, Gautam K, Tara K, Krishna P (2018) Recombinant human interferon regulatory factor-1 (IRF-1) protein expression and solubilisation study in Escherichia coli. Mol Biol Rep 45(5):1367–1374. https://doi.org/10.1007/s11033-018-4298-1
Saramago M, Bárria C, Arraiano CM, Domingues S (2015) Ribonucleases, antisense RNAs and the control of bacterial plasmids. Plasmid 78:26–36. https://doi.org/10.1016/j.plasmid2014.09.003
Schubert M, Edge RE, Lario P, Cook MA, Strynadka NC, Mackie GA, McIntosh LP (2004) Structural characterization of the RNase E S1 domain and identification of its oligonucleotide-binding and dimerization interfaces. J Mol Biol 341(1):37–54. https://doi.org/10.1016/j.jmb.2004.05.061
Singh RK, Jaiswal LK, Nayak T, Rawat RS, Kumar S, Rai SN, Gupta A (2022) Expression, Purification, and In Silico Characterization of Mycobacterium smegmatis Alternative Sigma Factor SigB. Dis Markers. https://doi.org/10.1155/2022/7475704
Sørensen HP, Mortensen KK (2005) Soluble expression of recombinant proteins in the cytoplasm of Escherichia coli. Microb Cell Fact 4(1):1–8. https://doi.org/10.1186/1475-2859-4-1
Su YC, Lim KP, Nathan S (2003) Bacterial expression of the scFv fragment of a recombinant antibody specific for Burkholderia pseudomallei exotoxin. BMB Rep 36(5):493–498. https://doi.org/10.5483/bmbrep.2003.36.5.493
Sun Y, Zhang J, Wang S (2015) Heterologous expression and efficient secretion of chitosanase from Microbacterium sp. Escherichia Coli Indian J Microbiol 55(2):194–199. https://doi.org/10.1007/s12088-014-0505-5
Trinquier A, Durand S, Braun F (1863) Regulation of RNA processing and degradation in bacteria. Biochim Biophys Acta Gene Regul Mech 5:194–505. https://doi.org/10.1016/j.bbagrm.2020.194505
Voss JE, Luisi BF, Hardwick SW (2014) Molecular recognition of RhlB and RNase D in the Caulobactercrescentus RNA degradosome. Nucleic Acids Res 42(21):13294–13305. https://doi.org/10.1093/nar/gku1134
Wlodawer A (2017) Stereochemistry and validation of macromolecular structures. Protein Crystallograph. https://doi.org/10.1007/978-1-4939-7000-1_24
Zeller ME, Csanadi A, Miczak A, Rose T, Bizebard T, Kaberdin VR (2007) Quaternary structure and biochemical properties of mycobacterial RNase E/G. Biochem J 403(1):207–215. https://doi.org/10.1042/BJ20061530
Zheng Z, Zhang MJ, Zhang G, Chen GQ (2004) Production of 3-hydroxydecanoic acid by recombinant Escherichia coli HB101 harboringpha G gene. Antonie Van Leeuwenhoek 85(2):93–101. https://doi.org/10.1023/B:ANTO.0000020275.23140.ca
Acknowledgements
LKJ, RKS and TN got the financial assistance from the Indian Council of Medical Research (ICMR) and University Grants Commission (UGC), New Delhi, respectively, as Senior Research Fellowship (SRF). We also acknowledge the central instrumentation facility of Interdisciplinary School of Life Sciences (ISLS), Banaras Hindu University, Varanasi, for providing us the instruments.
Author information
Authors and Affiliations
Contributions
L K J performed most of the experiments and mainly wrote the draft, R K S and T N helped in optimization of experimental conditions, V K S performed the bioinformatics study and A G designed the study and supervised the experiments and revised the final manuscript.
Corresponding author
Ethics declarations
Conflict of Interest
The authors declare that they have no conflict of interest.
Competing Interests
From the known literature, it is concluded that RNase E is the central enzyme involved in all aspect of RNA metabolism. There is very little information known about mycobacterial RNase E structure and function. Therefore, authors selected this gene for their study. Further, its regulation and interactors can be studied to show its possible role as drug target for pathogenic mycobacterial diseases.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Jaiswal, L.K., Singh, R.K., Nayak, T. et al. Heterologous Expression, Purification and Structural Characterization of Ribonuclease E from Mycobacterium smegmatis. Iran J Sci 47, 683–693 (2023). https://doi.org/10.1007/s40995-023-01467-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s40995-023-01467-x