Abstract
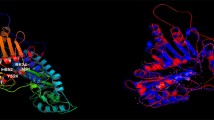
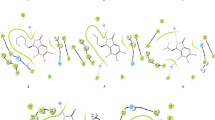
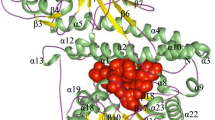
Cytidylate kinase (CMK) plays a central role in the phosphorylation of ATP to nucleoside diphosphates in the purine and pyrimidine pathway in Mycobacterium tuberculosis (MTB). The enzyme is specific to bacteria, fungi, and plants and it shares the same pathway with a DNA-directed RNA polymerase subunit beta, the known target of rifampicin (RIF). Therefore, it serves as an attractive alternative target to inhibit the purine and pyrimidine pathway in MTB. In this study, the novel inhibitors of CMK were identified using in silico approach. Homology modeling was used to build the three-dimensional (3D) structure of CMK using program implemented in Modeller 9.16 and based on the template (3R20) obtained from Mycobacterium smegmatis. Structural analysis of CMK showed that it has P-loop or core in the ATP binding domain, nucleoside monophosphate binding (NMP) site, and a LID domain. Inhibition of any of the residues that formed the nucleoside binding site or ATP binding site blocks the catalytic activity of the CMK. Eleven thousand four hundred and fifty-four compounds that had an affinity to bind to the CMK with minimum binding energies were obtained through virtual screening from Zinc and PubChem databases using PyRx 8.0. These compounds were further filtered for Lipinski rule of five, molecular docking analysis, and ADMET properties (absorption distribution, metabolism, excretion, and toxicity). Four compounds (ZINC95485891 = − 8.80 kcal/mol, ZINC95485880 = − 8.44 kcal/mol, ZINC95485900 = − 8.24 kcal/mol, and ZINC95486231 = − 8.20 kcal/mol) with good binding energies and desirable pharmacokinetic properties were selected. These compounds were further subjected to molecular dynamic (MD) simulation and molecular mechanics generalized Born and surface area (MM-GBSA) analyses. The results of the analyses showed that all the four ligands formed a stable complex and had good free binding energy after 50 ns MD simulation. Therefore, these compounds were considered as possible inhibitors of MTB after experimental validation.









Similar content being viewed by others
References
Al-Anazi KA, Al-Jasser AM, Alsaleh K (2014) Infections caused by MTB in recipients of hematopoietic stem cell transplantation. Front Oncol 4:231
Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25(17):3389–3402
Briozzo P, Golinelli-Pimpaneau B, Gilles AM, Gaucher JF, Burlacu-Miron S, Sakamoto H, Bârzu O (1998) Structures of Escherichia coli CMP kinase alone and in complex with CDP: a new fold of the nucleoside monophosphate binding domain and insights into cytosine nucleotide specificity. Structure 6(12):1517–1527
Caceres RA, Timmers LFSM, Vivan AL, Schneider CZ, Basso LA, De Azevedo WF, Santos DS (2008) Molecular modeling and dynamics studies of cytidylate kinase from Mycobacterium tuberculosis H37Rv. J Mol Model 14(5):427–434
Case DA, Berryman JT, Betz RM, Cerutti DS, Cheatham TE III, Darden TA, Duke RE et al (2015) AMBER 2015. University of California, San Francisco
Cheng F, Li W, Zhou Y, Shen J, Wu Z, Liu G, Tang Y (2012) AdmetSAR: a comprehensive source and free tool for assessment of chemical ADMET properties. J Chem Inf Model 52(11):3099–3105
Cloete R, Oppon E, Murungi E, Schubert WD, Christoffels A (2016) Resistance-related metabolic pathways for drug target identification in MTB. BMC Bioinform 17(1):75
Colovos C, Yeates TO (1993) Verification of protein structures: patterns of nonbonded atomic interactions. Protein Sci 2(9):1511–1519
DeLano WL (2002) The PyMOL user’s manual. DeLano Scientific, San Carlos, p 452
Esteves LS, Dalla Costa ER, Vasconcellos SEG, Vargas A, Junior SLMF, Halon ML, Rossetti MLR (2018) Genetic diversity of Mycobacterium tuberculosis isoniazid monoresistant and multidrug-resistant in Rio Grande do Sul, a tuberculosis high-burden state in Brazil. Tuberculosis 110:36–43
Genheden S, Ryde U (2015) The MM/PBSA and MM/GBSA methods to estimate ligand-binding affinities. Expert Opin Drug Discov 10(5):449–461
Ghasemi F, Zomorodipour A, Karkhane AA, Khorramizadeh MR (2016) In silico designing of hyper-glycosylated analogs for the human coagulation factor IX. J Mol Graph Model 68:39–47
La Motta C, Sartini S, Mugnaini L, Simorini F, Taliani S, Salerno S, Cantore M (2007) Pyrido[1,2-a]pyrimidine-4-one derivatives as a novel class of selective aldose reductase inhibitors exhibiting antioxidant activity. J Med Chem 50(20):4917–4927
Laskowski RA, Swindells MB (2011) LigPlot+: multiple ligand–protein interaction diagrams for drug discovery. J Chem Inf Model 51(10):2778–2786. https://doi.org/10.1021/ci200227u
Lipinski CA, Lombardo F, Dominy BW, Feeney PJ (2001) Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv Drug Deliv Rev 46(1–3):3–26
Lüthy R, Bowie JU, Eisenberg D (1992) Assessment of protein models with three-dimensional profiles. Nature 356(6364):83
Morris GM, Goodsell DS, Halliday RS, Huey R, Hart WE, Belew RK, Olson AJ (1998) Automated docking using a Lamarckian genetic algorithm and an empirical binding free energy function. J Comput Chem 19(14):1639–1662
Mukherjee G, Jayaram B (2013) Rapid identification of hit molecules for target proteins via physicochemical descriptors. Phys Chem Chem Phys 15(23):9107–9116
Palomino JC, Martin A (2014) Drug resistance mechanisms in MTB. Antibiotics 3(3):317–340
Pieper U, Webb BM, Barkan DT, Schneidman-Duhovny D, Schlessinger A, Braberg H, Datta RS (2010) ModBase, a database of annotated comparative protein structure models, and associated resources. Nucleic Acids Res 39(suppl_1):D465–D474
Rowland K (2012) Totally drug-resistant TB emerges in India. Nature 1:9797
Salomon-Ferrer R, Case DA, Walker RC (2013) An overview of the Amber biomolecular simulation package. Wiley Interdiscip Rev Comput Mol Sci 3(2):198–210
Thompson DJ, Higgins DG, Gibson TJ (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res 22(22):4673–4680
Thum C, Schneider CZ, Palma MS, Santos DS, Basso LA (2009) The Rv1712 Locus from MTB H37Rv codes for a functional CMP kinase that preferentially phosphorylates dCMP. J Bacteriol 191(8):2884–2887
Veber DF, Johnson SR, Cheng HY, Smith BR, Ward KW, Kopple KD (2002) Molecular properties that influence the oral bioavailability of drug candidates. J Med Chem 45(12):2615–2623
Wallace AC, Laskowski RA, Thornton JM (1996) Derivation of 3D coordinate templates for searching structural databases: application to Ser–His–Asp catalytic triads in the serine proteinases and lipases. Protein Sci 5(6):1001–1013
World Health Organization (2015) Digital health for the End TB Strategy: an agenda for action (no. WHO/HTM/TB/2015.21). World Health Organization. http://www.who.int/tb/areas-of-work/digital-health/Digital_health_EndTBstrategy.pdf. Accessed 8 Aug 2016
Yu L, Mack J, Hajduk PJ, Kakavas SJ, Saiki AY, Lerner CG, Olejniczak ET (2003) Solution structure and function of an essential CMP kinase of Streptococcus pneumoniae. Protein Sci 12(11):2613–2621
Zhang Y, Yew WW (2009) Mechanisms of drug resistance in MTB [State of the art series. Drug-resistant tuberculosis. Edited by CY. Chiang. Number 1 in the series]. Int J Tuberc Lung Dis 13(11):1320–1330
Zhang X, Zhang S, Hao F, Lai X, Yu H, Huang Y, Wang H (2005) Expression, purification and properties of shikimate dehydrogenase from MTB. J Biochem Mol Biol 38(5):624
Acknowledgements
The author of this paper greatly acknowledges Prof. B. Jayaram (Coordinator of the Supercomputing Facility for Bioinformatics and Computational Biology, IIT Delhi), Prof. Pawan Dhar (Jawaharlal Nehru University), Dr. Kalaiarasan P. (Jawaharlal Nehru University), and Mr. Shashank Shekhar (IIT Delhi) for their involvement and for providing facilities.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
I declare that i have no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Isa, M.A. Homology modeling and in silico toxicity assessment of potential inhibitors of cytidylate kinase from Mycobacterium tuberculosis. Netw Model Anal Health Inform Bioinforma 8, 16 (2019). https://doi.org/10.1007/s13721-019-0191-7
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1007/s13721-019-0191-7