Abstract
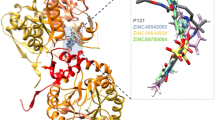
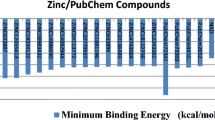
This study aimed to identify compounds that are capable of inhibiting polyphosphate kinase (PPK), the protein that shares a similar pathway with the target of isoniazid (INH) in the oxidative phosphorylation pathway of Mycobacterium tuberculosis (MTB). The three-dimensional structure of the PPK was predicted based on the homology modeling principle via Modeller9.16. Structural analysis revealed that the PPK has three active sites—metal ion-binding site (Arg431 and Arg461), ATP-binding site (Asn91, Tyr524, Arg624, and His652), and phosphohistidine intermediate active site (His491). The amino acids mentioned earlier play an essential role in the activity of the PPK, and their inhibition would block the function of the PPK. Ten thousand one hundred and six (10,106) ligands were obtained through virtual screening against Zinc database. These compounds were filtered by Lipinski rule of five and molecular docking analysis. A total of ten compounds with good AutoDock binding energy that varied between − 9.92 and − 8.01 kcal/mol which was lower than the binding energy of − 1.13 kcal/mol of Mg2+ (metallic cofactor) were selected. These compounds were further filtered for their pharmacokinetic and toxicity properties to further remove the compounds with unwanted properties. Three ligands—ZINC41125011, ZINC20318248, and ZINC20321877 with desired pharmacokinetic and toxicity properties were selected for molecular dynamic (MD) simulation and molecular generalized Born surface area (MM-GBSA) analysis. The result of the studies shows that all the three complexes are relatively stable in the binding site of the PPK, after 50 ns MD simulation. Therefore, these identified compounds are regarded as prospective inhibitors of MTB after positive experimental validation.









Similar content being viewed by others
References
Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25(17):3389–3402 (External Resources Pubmed/Medline (NLM) CrossRef (DOI) Chemical Abstracts Service (CAS) Cambridge Scientific Abstracts (CSA) ISI Web of Science)
Bernstein FC, Koetzle TF, Williams GJ, Meyer EF, Brice MD, Rodgers JR, Kennard O, Shimanouchi T, Tasumi M (1977) The Protein Data Bank. FEBS J 80(2):319–324
Case DA, Berryman JT, Betz RM, Cerutti DS, Cheatham TE III, Darden TA, Duke RE et al (2015) AMBER 2015. University of California, San Francisco
Centers for Disease Control and Prevention (2006) Emergence of MTB with extensive resistance to second-line drugs—worldwide, 2000–2004. MMWR Morb Mortal Wkly Rep 55(11):301
Chen C, Huang H, Wu CH (2017) Protein bioinformatics databases and resources. In: Wu CH, Arighi CN, Ross KE (eds) Protein bioinformatics. Humana Press, New York, pp 3–39
Cheng F, Li W, Zhou Y, Shen J, Wu Z, Liu G, Lee PW, Tang Y (2012) admetSAR: a comprehensive source and free tool for assessment of chemical ADMET properties. J Chem Inf Model 52:3099–3105
Cloete R, Oppon E, Murungi E, Schubert WD, Christoffels A (2016) Resistance related metabolic pathways for drug target identification in Mycobacterium tuberculosis. BMC Bioinform 17(1):75
Colovos C, Yeates TO (1993) Verification of protein structures: patterns of nonbonded atomic interactions. Protein Sci 2(9):1511–1519
DeLano WL (2002) The PyMOL user’s manual. DeLano Scientific, San Carlos, p 452
Fiser A, Do RKG (2000) Modeling of loops in protein structures. Protein Sci 9(9):1753–1773
Gasteiger J, Marsili M (1980) Iterative partial equalization of orbital electronegativity—a rapid access to atomic charges. Tetrahedron 36(22):3219–3228
Genheden S, Ryde U (2015) The MM/PBSA and MM/GBSA methods to estimate ligand-binding affinities. Expert Opin Drug Discov 10(5):449–461
Irwin JJ, Shoichet BK (2005) ZINC: a free database of commercially available compounds for virtual screening. J Chem Inf Model 45(1):177–182
Johnson R, Streicher EM, Louw GE, Warren RM, Van Helden PD, Victor TC (2006) Drug resistance in Mycobacterium tuberculosis. Curr Issues Mol Biol 8(2):97–112
Kim S, Thiessen PA, Bolton EE, Chen J, Fu G, Gindulyte A, Han L, He J, He S, Shoemaker BA, Wang J, Yu B, Zhang J, Bryant SH (2015) PubChem substance and compound databases. Nucleic Acids Res 44(D1):D1202–D1213
La Motta C, Sartini S, Mugnaini L, Simorini F, Taliani S, Salerno S, Marini AM, Da Settimo F, Lavecchia A, Novellino E, Cantore M, Failli P, Ciuffi M (2007) Pyrido [1, 2-a] pyrimidin-4-one derivatives as a novel class of selective aldose reductase inhibitors exhibiting antioxidant activity. J Med Chem 50(20):4917–4927
Laskowski RA, Swindells MB (2011) LigPlot+: multiple ligand-protein interaction diagrams for drug discovery. J Chem Inf Model 51(10):2778–2786
Lee AS, Teo AS, Wong SY (2001) Novel mutations in ndh in isoniazid-resistant Mycobacterium tuberculosis isolates. Antimicrob Agents Chemother 45(7):2157–2159
Lipinski CA, Lombardo F, Dominy BW, Feeney PJ (1997) Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv Drug Deliv Rev 23(1–3):3–25
Lüthy R, Bowie JU, Eisenberg D (1992) Assessment of protein models with three-dimensional profiles. Nature 356(6364):83
Martí-Renom MA, Stuart AC, Fiser A, Sánchez R, Melo F, Šali A (2000) Comparative protein structure modeling of genes and genomes. Annu Rev Biophys Biomol Struct 29(1):291–325
Morris GM, Goodsell DS, Halliday RS, Huey R, Hart WE, Belew RK, Olson AJ (1998) Automated docking using a Lamarckian genetic algorithm and an empirical binding free energy function. J Comput Chem 19(14):1639–1662
Mukherjee G, Jayaram B (2013) A rapid identification of hit molecules for target proteins via physico-chemical descriptors. Phys Chem Chem Phys 15(23):9107–9116
Murphy DJ, Brown JR (2007) Identification of gene targets against dormant phase Mycobacterium tuberculosis infections. BMC Infect Dis 7(1):84
Nightingale A, Antunes R, Alpi E, Bursteinas B, Gonzales L, Liu W, Luo J, Qi G, Turner E, Martin M (2017) The proteins API: accessing key integrated protein and genome information. Nucleic Acids Res 45(W1):W539–W544
Palomino JC, Martin A (2014) Drug resistance mechanisms in Mycobacterium tuberculosis. Antibiotics 3(3):317–340
Pieper U, Webb BM, Barkan DT, Schneidman-Duhovny D, Schlessinger A, Braberg H, Yang Z, Meng EC, Pettersen EF, Huang CC, Datta RS, Sampathkumar P, Madhusudhan MS, Sjölander K, Ferrin TE, Burley SK, Sali A (2010) ModBase, a database of annotated comparative protein structure models, and associated resources. Nucleic Acids Res 39(suppl_1):D465–D474
Rawat R, Whitty A, Tonge PJ (2003) The isoniazid-NAD adduct is a slow, tight-binding inhibitor of InhA, the Mycobacterium tuberculosis enoyl reductase: adduct affinity and drug resistance. Proc Natl Acad Sci 100(24):13881–13886
Rigi G, Nakhaei MVA, Eidipour H, Najimi A, Tajik F, Taher N, Yarahmadi K (2017) Virtual screening following rational drug design based approach for introducing new anti amyloid beta aggregation agent. Bioinformation 13(2):42
Sander T, Freyss J, von Korff M, Rufener C (2015) DataWarrior: an open-source program for chemistry aware data visualization and analysis. J Chem Inf Model 55(2):460–473
Thompson DJ, Higgins DG, Gibson TJ (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position specific gap penalties and weight matrix choice. Nucleic Acids Res 22(22):4673–4680
Vareldzis BP, Grosset J, de Kantor I, Crofton J, Laszlo A, Felten M, Raviglione MC, Kochi A (1994) Drug-resistant tuberculosis: laboratory issues. World Health Organization recommendations. Tuber Lung Dis 75:1–7
Veber DF, Johnson SR, Cheng HY, Smith BR, Ward KW, Kopple KD (2002) Molecular properties that influence the oral bioavailability of drug candidates. J Med Chem 45(12):2615–2623
Velayati AA, Masjedi MR, Farnia P, Tabarsi P, Ghanavi J, ZiaZarifi AH, Hoffner SE (2009) Emergence of new forms of totally drug-resistant tuberculosis bacilli: super extensively drug-resistant tuberculosis or totally drug-resistant strains in Iran. Chest 136(2):420–425
Wallace AC, Laskowski RA, Thornton JM (1996) Derivation of 3D coordinate templates for searching structural databases: application to Ser-His-Asp catalytic triads in the serine proteinases and lipases. Protein Sci 5(6):1001–1013
Webb B, Sali A (2014) Protein structure modeling with MODELLER. Method Mol Biol 1137:1–15
World Health Organization (2016) Global tuberculosis report. WHO, Geneva
Xu J, Yuan H, Ran T, Zhang Y, Liu H, Lu S, Xiong X, Xu A, Jiang Y, Lu T, Chen Y (2015) A selectivity study of sodium-dependent glucose cotransporter 2/sodium-dependent glucose cotransporter 1 inhibitors by molecular modeling. J Mol Recognit 28(8):467–479
Zhang Y, Heym B, Allen B, Young D, Cole S (1992) The catalase–peroxidase gene and isoniazid resistance of Mycobacterium tuberculosis. Nature 358(6387):591
Zhang X, Zhang S, Hao F, Lai X, Yu H, Huang Y, Wang H (2005) Expression, purification and properties of shikimate dehydrogenase from Mycobacterium tuberculosis. J Biochem Mol Biol 38(5):624
Acknowledgements
The authors of this paper are very much grateful to Prof. Pawan Dhar (Jawaharlal Nehru University), Prof. B. Jayaram (Indian Institute of Technology Delhi), Dr. Kalaiarasan P. (Jawaharlal Nehru University), and Mr. Shashank Shekhar (Indian Institute of Technology Delhi) for providing facilities.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
We declare that we have no conflict of Interest.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Alhaji Isa, M., Singh Majumdar, R. Computer-aided drug design based on comparative modeling, molecular docking and molecular dynamic simulation of Polyphosphate kinase (PPK) from Mycobacterium tuberculosis. J Proteins Proteom 10, 55–68 (2019). https://doi.org/10.1007/s42485-019-00006-w
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s42485-019-00006-w