Abstract
Glycyrrhiza glabra is one of the most important and well-known medicinal plants which produces various triterpene saponins such as glycyrrhizin. Beta-amyrin 11-oxidase (CYP88D6) plays a key role in engineering pathway of glycyrrhizin production and converts an intermediated beta-amyrin compound to glycyrrhizin. In this study, pBI121GUS-9:CYP88D6 construct was transferred to G. glabra using Agrobacterium rhizogene ATCC 15834. The quantitation of transgene was measured in putative transgenic hairy roots using qRT-PCR. The amount of glycyrrhizin production was measured by HPLC in transgenic hairy root lines. Gene expression analysis demonstrated that CYP88D6 was over-expressed only in one of transgenic hairy root lines and was reduced in two others. Beta-amyrin 24-hydroxylase (CYP93E6) was significantly expressed in one of the control hairy root lines. The amount of glycyrrhizin metabolite in over-expressed line was more than or similar to that of control hairy root lines. According to the obtained results, it would be recommended that multi-genes of glycyrrhizin biosynthetic pathway be transferred simultaneously to the hairy root in order to increase glycyrrhizin content.
Similar content being viewed by others
Introduction
Licorice is a popular medicinal plant used in many fields such as flavorings, medicaments, and cosmetics [1]. Glycyrrhizin is the most important triterpene saponin in roots and stolons of Glycyrrhiza (licorice) plant and is a conjugate of two molecules of glucuronic acid and glycyrrhetinic acid aglycone [2]. Glycyrrhizin demonstrates a wide range of pharmacological, antibacterial, hepato-protective [3, 4], antiulcer, anti-allergy [5], and anticancer [6,7,8] activities as well as antiviral function against various DNA and RNA viruses [5] including HIV [9] and severe acute respiratory syndrome-associated coronavirus [10]. Biosynthesis of triterpenoids in licorice starts with conversion of two molecules, namely farnesyl diphosphate (FDP) and squalene synthase enzyme (SQS) to squalene [11]. Then, oxidosqualene cyclases cause cyclization of 2,3-oxidosqualene to produce triterpenes and phytosterols [12]. Beta-amyrin synthase is one of the oxidosqualene cyclases, producing beta-amyrin which is responsible for biosynthesis of glycyrrhizin and soyasaponin triterpene saponins [13]. Oxidation of beta-amyrin at C-11 and C-30 positions produces glycyrrhizin, whereas oxidation at C-22 and C-24 positions leads to the production of soyasaponins [14,15,16]. Cytochrome P450s play critical roles in oxidative reactions during biosynthesis of diverse natural plant products. Beta-amyrin 11-oxidase (CYP88D6) is a cytochrome P450 which catalyzes two sequential oxidation steps of beta-amyrin at the position of C-11 to yield 11-oxo-beta-amyrin. Beta-amyrin 24-hydroxylase (CYP93E) is another P450 that catalyzes oxidation reaction of beta-amyrin at C-24 position and is a key initial enzyme in biosynthetic pathway of soyasaponins [15].
Hairy root cultures provide the potential for production of plant secondary metabolite and metabolic engineering by introducing or alteration the expression of specific metabolic pathway genes [17, 18]. These cultures possess genetic and biochemical stability, rapid growth rate, and high capability for synthesis of secondary metabolites at quantities comparable to the original plants [19]. A lot of researches have shown the increase in production of secondary metabolites using gene over-expression in transgenic hairy roots. The metabolic engineering approach increased production of flavonolignans through over-expressing petunia chalcone synthase (chsA) gene in hairy root cultures of Silybum marianum [20]. Over-expression of 7-O-acetyltransferase (SalAT) gene was employed as the target for metabolic engineering in Papaver bracteatum hairy root cultures, which led to the enhancement of morphinan alkaloid production [21]. There are a number of researches which have been successful in producing glycyrrhizin in G. glabra plant in cell and hairy root culture systems [22,23,24]. However, there is only one report on metabolic engineering of glycyrrhizin biosynthetic pathways in Glycyrrhiza plants, which uses gene transfer of squalene synthase 1 (SQS1) to enhance glycyrrhizin production in hairy root lines of Glycyrrhiza uralensis [11]. The cDNAs of CYP88D6 have been isolated and cloned from G. glabra [25], which provided the possibility to improve the study of glycyrrhizin pathway via metabolic engineering of G. glabra hairy root cultures.
In order to explore the potential of metabolic engineering of glycyrrhizin biosynthetic pathway, over-expression of CYP88D6 gene in hairy root cultures of G. glabra var. glabra was investigated. For this purpose, CYP88D6 was transferred into hairy root culture of G. glabra, followed by the expression of CYP88D6 and CYP93E6 genes, and subsequently, the amount of glycyrrhizin production in transgenic hairy roots was quantified.
Materials and Methods
Construction of CYP88D6 Expression Vector
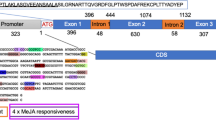
The full-length cDNA of CYP88D6 was isolated from seedlings of G. glabra [25]. The CYP88D6 coding sequence was digested with XbaI and BamHI, and was subcloned into pBI121GUS-9 plant vector (kindly provided by Dr. K. Esfahani, the National Institute of Genetic Engineering and Biotechnology, Iran) under control of a CaMV 35S promoter. Subsequently, pBI121GUS-9:CYP88D6 construct (Fig. 1) was introduced into Agrobacterium rhizogenes ATCC 15834 strain via electroporation. The A. rhizogenes having no binary vector was used for induction of the control hairy root. Prior to infection, mono-clone of ATCC 15834 was grown to mid-log phase (OD600 = 0.6) in liquid Luria–Bertani (LB) medium with 50 mg/L rifampicin and 100 mg/L kanamycin on a rotary shaker at 28 °C/110 rpm.
Hairy Root Transformation
Seeds of licorice (G. glabra L. var. glabra, Fabaceae) were provided by Pakan-Bazer Company (Isfahan, Iran). The seeds were disinfected using H2SO4 (98%) for 20 min and were subsequently washed with sterile water [23]. Then, they were cultured on solid MS medium [26] and incubated at 25 ± 2 °C for a photoperiod of 16 h light. Either 3-day-old seedlings or 3-week-old leaves were used as an explant for inoculation with A. rhizogenes containing the binary vector. The leaves were pricked with a sterile scalpel [23], and seedlings were cut off at root–hypocotyl transition site [27], which were then immersed in the overnight culture of bacteria suspension for 20 min and eventually blotted on a sterile filter paper. Some explants were inoculated with A. rhizogene devoid of binary vector which were served as controls. The inoculated explants were transferred to a solid MS medium containing 200 µM acetosyringone and were kept at 22 °C in darkness, during co-cultivation for 48 h [28]. In order to eliminate bacteria, the explants were washed in sterile distilled water and transferred to the fresh MS medium supplemented with 0.5 g/L cefotaxime. With emergence of hairy roots, they were selected on MS solid medium supplemented with 20 mg/L kanamycin. Then, 3–4 cm root pieces were transferred to MS liquid medium with 1% activated charcoal and were kept at 25 ± 2 °C on a rotary shaker (110 rpm) in darkness. The transformation frequency in leaves and seedlings was determined using a percentage of the number of explants inducing hairy roots to the total number of explants inoculated with A. rhizogene.
PCR Analysis of Putative Transgenics
Genomic DNA of hairy roots was extracted by CTAB method [29]. PCR was performed using A. rhizogenes VirD1 F 5′-ATGTCGCAAGGCAGTAAG-3′ and R 5′-CAAGGAGTCTTTCAGCATG-3′ as negative control to ensure the absence of bacteria in the results [30], rolB F 5′-TTAGGCTTCTTTCATTCGGTTTACTGCAGC-3′ and R 5′-ATGGATCCCAAATTGCTATTCCCCACGA-3′ to determine the integration of T-DNA from Ri plasmid into the roots [20], and CYP88D6 F 5′-ATGGAAGTACACTGGGTTTGC-3′ and R 5′-CTACGCACATGAAACCTTTATC-3′ to confirm the presence of CYP88D6 gene in transgenic hairy roots [25].
Gene Expression Analysis by Real-Time PCR
Total RNA was extracted from control and transgenic hairy roots using RNX-plus buffer (SinaClon, Iran) with some modifications. In order to remove genomic DNA contamination, total RNA was treated with RNase-free DNase I. 500 ng of total RNA from each sample was synthesized using oligo dT primer and RevertAid First Strand cDNA Synthesis Kit (Thermo Fisher). Real-time PCR was performed on BIO-RAD CFX96 real-time PCR system using 2X GreenHot Master Mix (BIORON GmbH, Germany). Expression analysis of transgenics was carried out using CYP88D6F (5′-TCCACGTCTTCATGGGCTCT-3′) and CYP88D6R (5′-CAACCGCCTTTCATCCACA-3′) as gene-specific primers for CYP88D6, as well as CYP93E6F (5′-AATGGGAGTGGGAGAAGAA-3′) and CYP93E6R (5′-AAATGGAACAAAACGAGGAAC-3′) as gene-specific primers for CYP93E6. ActinF (5′-GGCACCTCTCAACCCAAAAG-3′) and actinR (5′-GCTGACACCATCTCCAGAGT-3′) primers were used to quantify the house-keeping gene. PCR amplification was performed under the following conditions: initial denaturation at 95 °C for 5 min, 45 cycles of denaturation for 20 s at 95 °C, annealing for 20 s, extension at 72 °C for 20 s, and final extension at 72 °C for 7 min. Melting curves were analyzed so as to verify the reaction specificity. For expression assay, expression levels of the genes were calculated by comparing the cycle threshold value (Ct) for each gene to that of the reference gene using \(2^{{ -\Delta Ct }}\) method [31].
Chemical Analysis of Glycyrrhizin
40 mg of powdered hairy roots was extracted with 1 mL of methanol 80% (v/v) at 60 °C for 6 h. Extracts were centrifuged at 4000 rpm for 15 min at room temperature [32]. The supernatant was transferred to a new tube, and the solvent was evaporated at 30 °C for 5 h. Standard of glycyrrhizin (Sigma) along with residual extract was re-dissolved in 1 ml of methanol, followed by filtration through a 0.45 µm filter, and eventually the sample was subjected to HPLC. The separation was performed according to Sabbioni et al. method [33] using an isocratic separation on reversed-phase C18 column (150 × 4.6 mm i.d; 5 µm). Mobile phase consisted of methanol/acetonitrile/water/glacial acetic acid (35:34:30:1 by volume). A 20 µL aliquot of each extracted sample was analyzed by HPLC at room temperature. The applied flow rate was 1 mL/min, and detection wavelength was set at 254 nm.
Results and Discussion
Establishment of Transgenic Hairy Roots
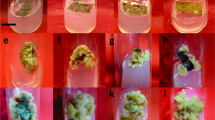
Direct generation of root emerged from wounded sites of leaf and seedling explants without callus formation on approximately 21–28 and 10 days after A. rhizogenes inoculation, respectively (Fig. 2a, b). The roots produced by the seedlings exhibited better growth which might be attributed to the normal growth of cotyledons and the presence of non-transgenic aerial parts. This plant was called as the composite due to the presence of transgenic root and non-transgenic aerial part. After growing, the roots were excised from the explants and were sub-cultured on the new MS medium with cefotaxime and kanamycin for transgenic harboring CYP88D6 transgene, and the medium having only cefotaxime for non-transgenic lines. Hairy roots produced by both methods showed morphological features such as rapid growth in hormone-free medium and high branching.
Induction of transgenic root of licorice using A. rhizogenes harboring pBI121GUS-9:CYP88D6 construct; a Hairy root induced on leaves explants; b composite plant culture with transgenic roots and non-transgenic aerial part; c rapid growth rate and branched roots in hormone-free MS solid medium; d hairy root culture in MS liquid medium with activated charcoal
After a while, surface of the Petri was covered with roots (Fig. 2c). The roots were transferred to MS liquid medium with activated charcoal (Fig. 2d). Due to the high release of phenolic compounds in liquid medium and its negative effects on the growth of roots, activated charcoal was used which had a favorable effect on root growth and also prevented browning of the tissue. Positive effects of activated charcoal on absorbance of toxic compounds including phenols have been previously reported in different plants [34]. Inoculation of plants with Agrobacterium rhizogenesis results in transfer, insertion, and expression of T-DNA from Ri plasmid in the plant genome, which leads to the expression of rolA, rolB, rolC, and rolD genes, as well as producing hairy roots in plants [35]. Another method for producing transgenic roots with A. rhizogenesis is the composite plant. The most important feature of this system is the reduced production time of transgenic root in plants. In addition to having hairy root culture properties, composite plant has other benefits such as the ability to grow in an ex vitro environment, which reduces the cost and time of production and simplifies the procedure [27]. Initially, the main reason for applying the composite plant method was to reduce the time of hairy roots production. On the other hand, the frequency of transgenic root production was 70% in composite plant method, whereas it was 20% in leaf explants. Plant transformation frequency differs significantly according to the source of the explant. The high frequency of hairy root production is related to tissue susceptibility and sensitivity to inoculation with A. rhizogenesis [36, 37].
Confirmation of Co-transformed Hairy Roots
PCR product using VirD1-specific primers showed a 441-bp band in bacterial plasmid, but no band was observed in hairy root samples (Fig. 3), indicating the absence of bacterial contamination in hairy roots. VirD1 gene is located outside the T-DNA region of the Ri plasmid, and hence, it is not theoretically transferred to the genome of the plant. The PCR product of rolB gene was detected in positive control plasmid and hairy root lines (Fig. 4).
In addition to verifying the hairy roots based on their morphology, a molecular method such as PCR was performed using rol gene-specific primers. The production of hairy roots in G. glabra, Gmelina arborea, S. marianum, and Plumbago rosea plants has been confirmed using rolB gene-specific primers [20, 23, 38, 39]. The integration of CYP88D6 gene into the hairy root was determined by PCR using CYP88D6-specific primers, showing a 1482-bp band in putative transgenic hairy roots, but not in control samples (Fig. 5). The results indicated that T-DNA from transformed A. rhizogenesis was successfully transferred into the three putative hairy root lines. These three lines had rolB and CYP88D6 transgenes which were named as D1, D2, and D3, while control hairy roots had only rolB transgene which were named HR1 and HR2. If in addition to the Ri plasmid, A. rhizogenesis has a binary vector, the process of co-transformation will occur with two T-DNAs [40]. Co-transformation in transgenic hairy roots has been reported in different of plants [37].
PCR product of rolB gene. M: 100-bp plus marker; 1: plasmid of A. rhizogenesis harboring pBI121GUS-9:CYP88D6 binary vector (positive control); 2, 4, 5: amplification of a 1482-bp band in transgenic hairy roots (harboring CYP88D6 transgene); 3: non-transgenic hairy roots (without CYP88D6 transgene, negative control)
The Expression of CYP88D6 and CYP93E6 in Hairy Roots
Expression levels of CYP88D6 and CYP93E6 were investigated in three transgenic and two control hairy root lines using qRT-PCR. CYP88D6 expression level showed a 6–12 fold induction in D1 transgenic hairy root line, and suppression in two other transgenic lines compared to control hairy roots (Fig. 6). Depending on the integration position of T-DNA into the host genome, different hairy root lines are generated [41]. The differences between various transgenes and control hairy root lines occur as a result of the position effects of transgenes, which lead to differences in gene expression and secondary metabolite production. Following rolB and CYP88D6 integration, there are numerous effects on expression of the transgenes. The main reason for over-expression of CYP88D6 in D1 transgenic line is the existence of constitutive and strong CaMV 35S promoter which results in high levels of transgene expression in hairy roots [42, 43]. The suppression of CYP88D6 expression in D2 and D3 transgenic lines compared to the control can be attributed to many factors such as position effect, gene copy number, gene rearrangement, physiological and epistasis effects, as well as methylation levels [44]. Probably, one of the main reasons for suppression of the gene in two transgenic lines is the existence of a similar copy of the CYP88D6 gene in genome of the hairy roots that resulted in co-suppression of internal and transgene copy of CYP88D6 gene. The CYP93E6 gene was expressed in HR1 line, but was not observed in other control and transgenic lines, which are related to the few amounts of transcript copies in these lines. CYP93E6 expression level showed a fourfold induction in HR1. This gene plays a role in the use of the common beta-amyrin precursor in competition with CYP88D6 gene. Different genes in the T-DNA region of Ri or binary vector are randomly integrated into genome of the hairy roots and alter the genetic content of the hosts, which can further exert a different regulatory effect on the genes involved in production pathway of secondary metabolites [41]. CYP93E6 was expressed in HR1, and the expression of CYP88D6 in this line was twofold more than that of HR2, which might be due to different genetic contents as a result of random integration of rol transgenes into the genome as well as host genetic alterations in HR1 and HR2.
Glycyrrhizin Production in Transgenic Hairy Roots
Content of glycyrrhizin varied among transgenic and control hairy roots (Fig. 7). D2 and D3 transgenic lines, having a reduced expression pattern of CYP88D6 compared to the controls, also displayed lower glycyrrhizin production. D1 which had a significant increase in expression of CYP88D6, demonstrated higher glycyrrhizin content in comparison with HR2, but lower than HR1. Failure to increase the amount of glycyrrhizin following over-expression of CYP88D6 in D1 transgenic line compared to HR1 could be due to the lack of downstream gene expression, and hence protein down-regulation. It has been shown that when SQS1 isolated from Glycyrrhiza uralensis was transferred using Ri under CaMV 35S promoter to licorice, it produced three transgenic lines, two of which showed more glycyrrhizin content, and a lower production was observed in one of transgenic lines in comparison with control hairy root [11]. The putrescine N-methyltransferase (pmt) over-expression in hairy root of two Hyoscyamus species led to a higher level of hyoscyamine and total alkaloid content, but no linear relationship was showed between these metabolites. This could be attributed to the role of other genes contributing to the biosynthetic pathway of tropane alkaloids and consequently its complex regulation [45].
Generally, increasing the rate-limiting enzymes in biosynthetic pathways results in a rise in the amount of the desired metabolite; however, in some biosynthetic pathways there is more than one rate-limiting enzyme and consequently, over-expression of an enzyme cannot increase the production of the target metabolite. In such cases, there is a multi-genes transfer mechanism or a transcription factor affecting multiple genes [46]. It has also been observed that over-expression of pmt alone in Atropa belladonna did not affect tropane alkaloid production in transgenic hairy roots [47].
Conclusions
Metabolic engineering approach via over-expression of single gene may lead to an increase in gene expression involved in metabolic pathway, and hence accumulation of the final product. The present study is the first report regarding transfer of beta-amyrin 11-oxidase gene to G. glabra using hairy root system that is an alternative way to improve glycyrrhizin production. It seems that the pathway of glycyrrhizin production is very complicated and needs to examine the entire pathway in terms of the genes expression, enzymes activity, and the amount of intermediate metabolites production. Although the amount of glycyrrhizin increased in one of transgenic lines, in order to increase glycyrrhizin content in most of the lines, transfer of multi-genes of the biosynthetic pathway along with up-stream genes might be necessary so as to increase the primary precursor.
References
Hanrahan, C. (2001). Gale encyclopedia of alternative medicine: licorice. Thomson Gale: Farmington Hills.
Hayashi, H. (2009). Molecular biology of secondary metabolism: Case study for Glycyrrhiza plants. In A. Kirakosyan & P. B. Kaufman (Eds.), Recent advances in plant biotechnology (pp. 89–103). New York: Springer.
Chan, H. T., Chan, C., & Ho, J. W. (2003). Inhibition of glycyrrhizic acid on aflatoxin B1-induced cytotoxicity in hepatoma cells. Toxicology, 188, 211–217.
Jeong, H. G., You, H. J., Park, S. J., Moon, A. R., Chung, Y. C., Kang, S. K., et al. (2002). Hepatoprotective effects of 18β-glycyrrhetinic acid on carbon tetrachloride-induced liver injury: Inhibition of cytochrome P450 2E1 expression. Pharmacological Research, 46, 221–227.
Park, H. Y., Park, S. H., Yoon, H. K., Han, M. J., & Kim, D. H. (2004). Anti-allergic activity of 18β-glycyrrhetinic acid-3-O-β-d-glucuronide. Archives of Pharmacalogy Research, 27, 57–60.
Fiore, C., Eisenhut, M., Krausse, R., Ragazzi, E., Pellati, D., Armanini, D., et al. (2008). Antiviral effects of Glycyrrhiza species. Physiotherapy Research, 22, 141–148.
Salvi, M., Fiore, C., Armanini, D., & Toninello, A. (2003). Glycyrrhetinic acid-induced permeability transition in rat liver mitochondria. Biochemistry and Pharmacology, 66, 2375–2379.
Yoon, G., Do Jung, Y., & Cheon, S. H. (2005). Cytotoxic allyl retrochalcone from the roots of Glycyrrhiza inflata. Chemical & Pharmaceutical Bulletin, 53, 694–695.
De Clercq, E. (2000). Current lead natural products for the chemotherapy of human immunodeficiency virus (HIV) infection. Medicinal Research Reviews, 20, 323–349.
Cinatl, J., Morgenstern, B., Bauer, G., Chandra, P., Rabenau, H., & Doerr, H. (2003). Glycyrrhizin, an active component of liquorice roots, and replication of SARS-associated coronavirus. The Lancet, 361, 2045–2046.
Lu, H. Y., Liu, J. M., Zhang, H. C., Yin, T., & Gao, S. L. (2008). Ri-mediated transformation of Glycyrrhiza uralensis with a squalene synthase gene (GuSQS1) for production of glycyrrhizin. Plant Molecular Biology Reporter, 26, 1–11.
Haralampidis, K., Bryan, G., Qi, X., Papadopoulou, K., Bakht, S., Melton, R., et al. (2001). A new class of oxidosqualene cyclases directs synthesis of antimicrobial phytoprotectants in monocots. Proceedings of the National Academy of Sciences, 98, 13431–13436.
Hayashi, H., Huang, P., Kirakosyan, A., Inoue, K., Hiraoka, N., Ikeshiro, Y., et al. (2001). Cloning and characterization of a cDNA encoding β-amyrin synthase involved in glycyrrhizin and soyasaponin biosyntheses in licorice. Biological and Pharmaceutical Bulletin, 24, 912–916.
Hayashi, H., Hiraoka, N., Ikeshiro, Y., Kushiro, T., Morita, M., Shibuya, M., et al. (2000). Molecular cloning and characterization of a cDNA for Glycyrrhiza glabra cycloartenol synthase. Biological and Pharmaceutical Bulletin, 23, 231–234.
Seki, H., Ohyama, K., Sawai, S., Mizutani, M., Ohnishi, T., Sudo, H., et al. (2008). Licorice β-amyrin 11-oxidase, a cytochrome P450 with a key role in the biosynthesis of the triterpene sweetener glycyrrhizin. Proceedings of the National Academy of Sciences, 105, 14204–14209.
Hayashi, H., & Sudo, H. (2009). Economic importance of licorice. Plant Biotechnology, 26, 101–104.
Georgiev, M. I., Ludwig-Muller, J., & Bley, T. (2010). Hairy root culture: Copying nature in new bioprocesses. In R. Arora (Ed.), Medicinal plant biotechnology (pp. 156–175). Oxon: CAB International.
Ono, N. N., & Tian, L. (2011). The multiplicity of hairy root cultures: Prolific possibilities. Plant Science, 180, 439–446.
Sevon, N., & Oksman-Caldentey, K. M. (2002). Agrobacterium rhizogenes-mediated transformation: Root cultures as a source of alkaloids. Planta Medica, 68, 859–868.
Rahnama, H., Razi, Z., Dadgar, M. N., & Hasanloo, T. (2013). Enhanced production of flavonolignans in hairy root cultures of Silybum marianum by over-expression of chalcone synthase gene. Journal of Plant Biochemistry and Biotechnology, 22, 138–143.
Sharafi, A., Sohi, H. H., Mousavi, A., Azadi, P., Khalifani, B. H., & Razavi, K. (2013). Metabolic engineering of morphinan alkaloids by over-expression of codeinone reductase in transgenic hairy roots of Papaver bracteatum, the Iranian poppy. Biotechnology Letters, 35, 445–453.
Li, Y. L., Yang, Y., Fu, C. H., & Yu, L. J. (2012). Production of glycyrrhizin in cell suspension of Glycyrrhiza inflata Batalin cultured in bioreactor. Biotechnology and Biotechnological Equipment, 26, 3231–3235.
Shirazi, Z., Piri, K., Asl, A. M., & Hasanloo, T. (2012). Glycyrrhizin and isoliquiritigenin production by hairy root culture of Glycyrrhiza glabra. Journal of Medicinal Plants Research, 6, 4640–4646.
Mousa, N. A., Siaguru, P., Wiryowidagdo, S., & Wagih, M. (2007). Establishment of regenerative callus and cell suspension system of licorice (Glycyrrhiza glabra) for the production of the sweetener glycyrrhizinin vitro. Sugar Tech, 9, 72–82.
Shirazi, Z., Aalami, A., Tohidfar, M., & Sohani, M. M. (2016). Isolation, cloning and bioinformatics analysis of beta-amyrin 11-oxidase coding sequence from licorice. Plant Omics, 9, 165.
Murashige, T., & Skoog, F. (1962). A revised medium for rapid growth and bio assays with tobacco tissue cultures. Physiologia Plantarum, 15, 473–497.
Collier, R., Fuchs, B., Walter, N., Kevin, W., & Taylor, C. G. (2005). Ex vitro composite plants: An inexpensive. Rapid method for root biology. The Plant Journal, 43, 449–457.
Zhang, H. C., Liu, J. M., Chen, H. M., Gao, C. C., Lu, H. Y., Zhou, H., et al. (2011). Up-regulation of licochalcone A biosynthesis and secretion by Tween 80 in hairy root cultures of Glycyrrhiza uralensis Fisch. Molecular Biotechnology, 47, 50–56.
Cai, D., Kleine, M., Kifle, S., Harloff, H. J., Sandal, N. N., Marcker, K. A., et al. (1997). Positional cloning of a gene for nematode resistance in sugar beet. Science, 275, 832–834.
Vojin, T., Snezana, M., Aleksandar, C., Marija, P., Milana, T., & Dragana, A. (2014). Production of hairy root cultures of lettuce (Lactuca sativa). Open Life Sciences, 9, 1196–1205.
Pfaffl, M. W. (2001). A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Research, 29, e45.
Hayashi, H., Fukui, H., & Tabata, M. (1988). Examination of triterpenoids produced by callus and cell suspension cultures of Glycyrrhiza glabra. Plant Cell Reports, 7, 508–511.
Sabbioni, C., Ferranti, A., Bugamelli, F., Forti, G. C., & Raggi, M. A. (2006). Simultaneous HPLC analysis, with isocratic elution, of glycyrrhizin and glycyrrhetic acid in liquorice roots and confectionery products. Phytochemical Analysis, 17, 25–31.
Fridborg, G., Pedersen, M., Landstrom, L. E., & Eriksson, T. (1978). The effect of activated charcoal on tissue cultures: Adsorption of metabolites inhibiting morphogenesis. Physiologia Plantarum, 43, 104–106.
Christey, M. C., & Braun, R. H. (2004). Production of hairy root cultures and transgenic plants by Agrobacterium rhizogenes-mediated transformation (pp. 47–60). Methods and Protocols: Transgenic Plants.
Shi, H. P., & Kintzios, S. (2003). Genetic transformation of Pueraria phaseoloides with Agrobacterium rhizogenes and puerarin production in hairy roots. Plant Cell Reports, 21, 1103–1143.
Alpizar, E., Decham, E., Espeout, S., Royer, M., Lecouls, A. C., & Nicole, M. (2006). Efficient production of Agrobacterium rhizogenes-transformed roots and composite plants for studying gene expression in coffee roots. Plant Cell Reports, 25, 959–967.
Dhakulkar, S., Ganapathi, T., Bhargava, S., & Bapat, V. (2005). Induction of hairy roots in Gmelina arborea Roxb. and production of verbascoside in hairy roots. Plant Science, 169, 812–818.
Jose, B., Pillai, D. B., & Satheeshkumar, K. (2016). In vitro cultivation of hairy roots of Plumbago rosea L. in a customized reaction kettle for the production of plumbagin––An anticancer compound. Industrial Crops and Products, 87, 89–95.
Ilina, E. L., Logachov, A. A., Laplaze, L., Demchenko, N. P., Pawlowski, K., & Demchenko, K. N. (2012). Composite Cucurbita pepo plants with transgenic roots as a tool to study root development. Annals of Botany, 110, 479–489.
Hu, Z. B., & Du, M. (2006). Hairy root and its application in plant genetic engineering. Journal of Integrative Plant Biology, 48, 121–127.
Chattopadhyay, T., Roy, S., Mitra, A., & Maiti, M. K. (2011). Development of a transgenic hairy root system in jute (Corchorus capsularis L.) with gusA reporter gene through Agrobacterium rhizogenes mediated co-transformation. Plant Cell Reports, 30, 485–493.
Kim, S. R., Sim, J. S., Ajjappala, H., Kim, Y. H., & Hahn, B. S. (2012). Expression and large-scale production of the biochemically active human tissue-plasminogen activator in hairy roots of Oriental melon (Cucumis melo). Journal of Bioscience and Bioengineering, 113, 106–111.
Wilson, K. J., Hughes, S., & Jefferson, R. (1992). The Escherichia coli gus operon: Induction and expression of the gus operon in E. coli and the occurrence and use of GUS in other bacteria. In S. R. Gallagher (Ed.), GUS protocols: Using the GUS gene as a reporter of gene expression (pp. 125–145). San Diego: Harcourt Brace Jovanovich.
Dehghan, E., Reed, D. W., Covello, P. S., Hasanpour, Z., Palazon, J., Oksman-Caldentey, K. M., et al. (2017). Genetically engineered hairy root cultures of Hyoscyamus senecionis and H. muticus: Ploidy as a promising parameter in the metabolic engineering of tropane alkaloids. Plant Cell Reports, 36, 1615–1626.
Chandra, S. (2012). Natural plant genetic engineer Agrobacterium rhizogenes: Role of T-DNA in plant secondary metabolism. Biotechnology Letters, 34, 407–415.
Sato, F., Hashimoto, T., Hachiya, A., Tamura, K. I., Choi, K. B., & Morishige, T. (2001). Metabolic engineering of plant alkaloid biosynthesis. Proceedings of the National Academy of Sciences, 98, 367–372.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Shirazi, Z., Aalami, A., Tohidfar, M. et al. Metabolic Engineering of Glycyrrhizin Pathway by Over-Expression of Beta-amyrin 11-Oxidase in Transgenic Roots of Glycyrrhiza glabra. Mol Biotechnol 60, 412–419 (2018). https://doi.org/10.1007/s12033-018-0082-7
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12033-018-0082-7