Abstract
Key message
Through high-throughput sequencing, we compared the relative expression levels of miRNA in three full-sib Populus triploid populations with that in their parents and one diploid hybrid population. We found similar numbers of miRNAs differentially expressed between the parents and the four progeny hybrid populations. In addition, unbalanced parental expression level dominance of miRNAs were found in the three allotriploid and interspecific hybrid populations, which may reprogram gene expression networks and contribute to the growth of Populus hybrids. These results indicated that hybridization has a great impact on the miRNA expression variation in the newly synthesized Populus triploid and diploid hybrid populations. However, we also found no significant differences in miRNA expression among one diploid and three triploid hybrid populations, hinting that miRNA abundances do not increase with the genome content. No dosage effect of miRNA expression could lead to dosage-dependent negative effects on target genes and their downstream pathway in polyploids. We speculate that polyploids may gain advantages from the slight decrease in miRNA regulation, suggesting an important molecular mechanism of polyploid advantage.
Abstract
Hybridization with three types of induced 2n gametes transmitted different parental heterozygosities has been proven as an efficient method for Populus triploid production. Several researches have shown that miRNA could be non-additively expressed in allopolyploids. However, it is still unclear whether the non-additively expressed miRNAs result from the effect of hybridization or polyploidization, and whether a dose response to the additional genomic content exists for the expression of miRNA. Toward this end, through high-throughput sequencing, we compared the expression levels of miRNA in three full-sib Populus triploid populations with that in their parents and one interspecific hybrid population. We found similar numbers of miRNAs differentially expressed between the parents and the four progeny hybrid populations. Unbalanced parental expression level dominance of miRNAs were found in the three triploid and diploid hybrid populations, which may reprogram gene expression networks and affect the growth of Populus hybrids. These results indicated that hybridization has a great impact on the miRNA expression variation in the newly synthesized Populus triploid and diploid hybrid populations. However, we also found no significant differences in miRNA expression among the three triploid populations and the diploid hybrid population. No dosage effect of miRNA expression could lead to dosage-dependent negative effects on target genes and their downstream pathway in polyploids. We speculate that polyploids may gain advantages from the decrease in miRNA negative regulation, suggesting an important molecular mechanism of polyploid advantage.






Similar content being viewed by others
References
Adams KL, Wendel JF (2005) Polyploidy and genome evolution in plants. Curr Opin Plant Biol 8:135–141
An J, Lai J, Lehman M, Nelson C (2013) miRDeep*: an integrated application tool for miRNA identification from RNA sequencing data. Nucl Acids Res 41:727–737
Ashburner M, Ball CA, Blake JA et al (2000) Gene ontology: tool for the unification of biology. The gene ontology consortium. Nat Genet 25(1):25–29
Auger DL, Anjali Dogra G, Ream TS, Akio K, Coe EH, Birchler JA (2005) Nonadditive gene expression in diploid and triploid hybrids of maize. Genetics 169:389–397
Bardil A, Almeida JDD, Combes MC, Lashermes P, Bertrand B (2011) Genomic expression dominance in the natural allopolyploid coffea arabica is massively affected by growth temperature. New Phytol 192:760–774
Bartel DP (2004) MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116:281–297
Benjamini Y, Dan D, Elmer G, Kafkafi N, Golani I (2001) Controlling the false discovery rate in behavior genetics research. Behav Brain Res 125:279–284
Chagué V, Just J, Mestiri I, Balzergue S, Tanguy AM, Huneau C et al (2010) Genome-wide gene expression changes in genetically stable synthetic and natural wheat allohexaploids. New Phytol 187:1181–1194
Chelaifa H, Monnier A, Ainouche M (2010) Transcriptomic changes following recent natural hybridization and allopolyploidy in the salt marsh species Spartina x townsendii and Spartina anglica (Poaceae). New Phytol 186:161–174
Chen ZJ (2007) Genetic and epigenetic mechanisms for gene expression and phenotypic variation in plant polyploids. Annu Rev Plant Biol 58:377–406
Chen ZJ (2010) Molecular mechanisms of polyploidy and hybrid vigor. Trends Plant Sci 15(2):57
Cheng SP, Yang J, Liao T, Zhu XH, Suo YJ, Zhang PD, Wang J, Kang XY (2015) Transcriptomic changes following synthesis of a Populus full-sib diploid and allotriploid population with different heterozygosities driven by three types of 2n female gamete. Plant Mol Biol 89:493–510
Dai X, Zhao X (2011) psRNATarget: a plant small RNA target analysis server. Nucl Acids Res 39:W155–W159
Dillies MA, Rau A, Aubert J, Hennequet-Antier C, Jeanmougin M, Servant N et al (2013) A comprehensive evaluation of normalization methods for illumina high-throughput RNA sequencing data analysis. Brief Bioinform 14:671–683
Dong CB, Mao JF, Suo YJ, Shi L, Wang J, Zhang PD, Kang XY (2014) A strategy for characterization of persistent heteroduplex dna in higher plants. Plant J 80:282–291
Dong CB, Suo YJ, Wang J, Kang XY (2015) Analysis of transmission of heterozygosity by 2n gametes in Populus (Salicaceae). Tree Genet Genomes 11:1–7
Duan H, Lu X, Lian CL, An Y, Xia XL, Yin WL (2016) Genome-wide analysis of microRNA responses to the phytohormone abscisic acid in Populus euphratica. Front Plant Sci 7:449
Dubcovsky J, Dvorak J (2007) Genome plasticity a key factor in the success of polyploid wheat under domestication. Science 316:1862–1866
Flagel L, Wendel JF (2010) Evolutionary rate variation, genomic dominance and duplicate gene expression evolution during allotetraploid cotton speciation. New Phytol 186:184–193
Flagel L, Udall J, Dan N, Wendel JF (2008) Duplicate gene expression in allopolyploid Gossypium reveals two temporally distinct phases of expression evolution. BMC Biol 6:1–9
Gaeta RT, Yoo SY, Pires JC, Doerge RW, Chen ZJ, Osborn TC (2009) Analysis of gene expression in resynthesized Brassica napus allopolyploids using arabidopsis 70mer oligo microarrays. PLoS ONE 4:e4760
Grover CE, Gallagher JP, Szadkowski EP, Yoo MJ, Flagel LE, Wendel JF (2012) Homoeolog expression bias and expression level dominance in allopolyploids. New Phytol 196:966–971
Ha M, Lu J, Tian L, Ramachandran V, Kasschau KD, Chapman EJ, Carrington JC, Chen XM, Wang XJ, Chen ZJ (2009) Small RNAs serve as a genetic buffer against genomic shock in Arabidopsis interspecific hybrids and allopolyploids. PNAS 106:17835–17840
Hegarty MJ, Barker GL, Wilson ID, Abbott RJ, Edwards KJ, Hiscock SJ (2006) Transcriptome shock after interspecific hybridization in Senecio is ameliorated by genome duplication. Curr Biol 16:1652–1659
Ilut DC, Coate JE, Luciano AK, Owens TG, May GD, Farmer A, Doyle JJ (2012) A comparative transcriptomic study of an allotetraploid and its diploid progenitors illustrates the unique advantages and challenges of rna-seq in plant species. Am J Bot 99(2):383–396
Jackson S, Chen ZJ (2010) Genomic and expression plasticity of polyploidy. Curr Opin Plant Biol 13:153–159
Kenan-Eichler M, Leshkowitz D, Tal L, Noor E, Melamed-Bessudo C, Feldman M et al (2011) Wheat hybridization and polyploidization results in deregulation of small RNAs. Genetics 188:263–272
Kidner CA, Martienssen RA (2003) Macro effects of microRNAs in plants. Trends Genet 19:13–16
Li H, Durbin R (2009) Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics 25(14):1754–1760
Li A, Liu D, Wu J, Zhao X, Hao M, Geng S, Yan J, Jiang X, Zhang L, Wu J et al (2014) mRNA and small RNA transcriptomes reveal insights into dynamic homoeolog regulation of allopolyploid heterosis in nascent hexaploid wheat. Plant Cell 26:1878–1900
Liao T, Cheng SP, Zhu XH, Min Y, Kang XY (2016) Effects of triploid status on growth, photosynthesis, and leaf area in Populus. Trees 30(4):1–11
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(-delta delta C(T)) method. Methods 25:402–408
Lu M, Zhang PD, Kang XY (2013) Induction of 2n female gametes in Populus adenopoda Maxim by high temperature exposure during female gametophyte development. Breed Sci 63:96–103
Mason A, Batley J (2015) Creating new interspecific hybrid and polyploid crops. Trends Biotechnol 33:436–441
Osborn TC, Pires JC, Birchler JA, Auger DL, Chen ZJ, Lee HS et al (2003) Understanding mechanisms of novel gene expression in polyploids. Trends Genet 19:141–147
Paterson AH, Wendel JF, Gundlach H, Guo H, Jenkins J, Jin DC et al (2012) Repeated polyploidization of Gossypium genomes and the evolution of spinnable cotton fibres. Nature 492:423–427
Rapp RA, Udall JA, Wendel JF (2009) Genomic expression dominance in allopolyploids. BMC Biol 7:1–10
Reinhart BJ, Weinstein EG, Rhoades MW, Bartel B, Bartel DP (2002) MicroRNAs in plants. Genes Dev 16:1616–1626
Smyth GK (2004) Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol 3:1–28
Suo YJ, Dong CB, Kang XY (2015) Inheritance and variation of cytosine methylation in three Populus allotriploid populations with different heterozygosity. PLoS ONE 10:e0126491
Vaucheret H (2006) Post-transcriptional small RNA pathways in plants: mechanisms and regulations. Genes Dev 20:759–771
Voinnet O (2009) Origin, biogenesis, and activity of plant microRNAs. Cell 136:669–687
Wang JL, Tian L, Lee HS, Wei NE, Jiang HM, Watson B et al (2006) Genome wide nonadditive gene regulation in Arabidopsis allotetraploids. Genetics 172:507–517
Wang J, Kang XY, Li DL, Chen HW, Zhang PD (2010) Induction of diploid eggs with colchicines during embryo sac development in Populus. Silvae. Genetica 59:40
Wang J, Kang XY, Li DL (2012a) High temperature-induced triploid production during embryo sac development in Populus. Silvae Genet 61:85–93
Wang J, Li DL, Kang XY (2012b) Induction of unreduced megaspores with high temperature during megasporogenesis in Populus. Ann For Sci 69:59–67
Wendel JF (2000) Genome evolution in polyploids. Plant Mol Biol 42:225–249
Zhang Q, Zhang ZY, Lin SZ, Lin YZ (2005) Resistance of transgenic hybrid triploids in Populus tomentosa Carr. against 3 species of lepidopterans following two winter dormancies conferred by high level expression of cowpea trypsin inhibitor gene. Silvae Genet 54:108–116
Zhang PD, Wu F, Kang XY (2011) Differences in fiber properties and basic wood density in triploid hybrids of Populus tomentosa. Adv Mater Res 236–238:1442–1452
Zhu ZT, Kang XY, Zhang ZY (1998) Studies on selection of natural triploids of Populus tomentosa. Sci Silvae Sin 34:22–31
Acknowledgements
This work was financially supported by National Natural Science Foundation of China (31530012), Special Fund for Forest Scientific Research in the Public Welfare (201404113), National Natural Science Foundation of China (31470667). We thank Dr. Hui Duan for assistance with technical advice, Dai Chen, Bo Zhang and Yang Yang (Novel Bioinformatics Ltd., Co, Shanghai, China) for assistance in bioinformatics analysis.
Author information
Authors and Affiliations
Contributions
YS and XK conceived the project; YS and SC designed experiments; YS, YM and YW performed most of the experiments; YS and CD analysed data; YS wrote the paper; CD reviewed the manuscript; all authors commented on the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
No conflict of interest declared.
Electronic supplementary material
Below is the link to the electronic supplementary material.
11103_2017_627_MOESM1_ESM.tif
The developmental processes of Populus triploid resulting from the inhibition of first division restitution, second division restitution, and post-meiotic restitution (TIF 2133 KB)
11103_2017_627_MOESM2_ESM.tif
GO term enrichment analysis of maternal-specific miRNAs (FDR <0.05). BP, biological process; MF, molecular function; CC, cellular component (TIF 1420 KB)
11103_2017_627_MOESM3_ESM.tif
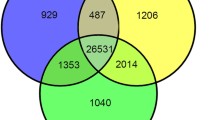
Venn diagram of ELD miRNAs in three allotriploid populations and one diploid F1 population. a, maternal ELD miRNAs; b, paternal ELD miRNAs (TIF 1546 KB)
Rights and permissions
About this article
Cite this article
Suo, Y., Min, Y., Dong, C. et al. MicroRNA expression changes following synthesis of three full-sib Populus triploid populations with different heterozygosities. Plant Mol Biol 95, 215–225 (2017). https://doi.org/10.1007/s11103-017-0627-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11103-017-0627-3