Abstract
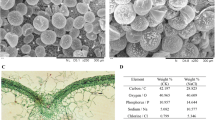
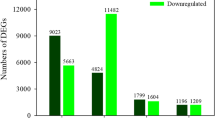
Limonium bicolor, a typical recretohalophyte that lives in saline environments, excretes excessive salt to the environment through epidermal salt glands to avoid salt stress. The aim of this study was to screen for L. bicolor genes involved in salt secretion by high-throughput RNA sequencing. We established the experimental procedure of salt secretion using detached mature leaves, in which the optimal salt concentration was determined as 200 mM NaCl. The detached salt secretion system combined with Illumina deep sequencing were applied. In total, 27,311 genes were annotated using an L. bicolor database, and 2040 of these genes were differentially expressed, of which 744 were up-regulated and 1260 were down-regulated with the NaCl versus the control treatment. A gene ontology enrichment analysis indicated that genes related to ion transport, vesicles, reactive oxygen species scavenging, the abscisic acid-dependent signaling pathway and transcription factors were found to be highly expressed under NaCl treatment. We found that 102 of these genes were likely to be involved in salt secretion, which was confirmed using salt-secretion mutants. The present study identifies the candidate genes in the L. bicolor salt gland that are highly associated with salt secretion. In addition, a salt-transporting pathway is presented to explain how Na+ is excreted by the salt gland in L. bicolor. These findings will shed light on the molecular mechanism of salt secretion from the salt glands of plants.







Similar content being viewed by others
References
Apse MP, Aharon GS, Snedden WA, Blumwald E (1999) Salt tolerance conferred by overexpression of a vacuolar Na+/H+ antiport in Arabidopsis. Science 285:1256–1258
Arisz WH, Camphuis IJ, Heikens H, Av Tooren (1955) The secretion of the salt glands of Limonium latifolium Ktze. Acta Bot Neerl 4(3):322–338
Arrial RT, Togawa RC, Brigido MM (2009) Screening non-coding RNAs in transcriptomes from neglected species using PORTRAIT: case study of the pathogenic fungus Paracoccidioides brasiliensis. BMC Bioinform 10:239
Barkla BJ, Zingarelli L, Blumwald E, Smith JAC (1995) Tonoplast Na+/H+ antiport activity and its energization by the vacuolar H+-ATPase in the halophytic plant Mesembryanthemum crystallinum L. Plant Physiol 109:549–556
Barkla BJ, Vera-Estrella R, Camacho-Emiterio J, Pantoja O (2002) Na+/H+ exchange in the halophyte Mesembryanthemum crystallinum is associated with cellular sites of Na+ storage. Funct Plant Biol 29:1017–1024
Breckle SW (1995) How do halophytes overcome salinity? In: Khan MA, Ungar IA (eds) Biology of salt tolerant plants. Book Graffers, Chelsea, pp 199–213
Chen SH, Guo SL, Wang ZL, Zhao JQ, Zhao YX, Zhang H (2007) Expressed sequence tags from the halophyte Limonium sinense: full length research article. Mitochondrial DNA 18:61–67
Cominelli E, Galbiati M, Vavasseur A, Conti L, Sala T, Vuylsteke M, Leonhardt N, Dellaporta SL, Tonelli C (2005) A guard-cell-specific MYB transcription factor regulates stomatal movements and plant drought tolerance. Curr Biol 15:1196–1200
Cvrčková F, Bezvoda R, Žárský V (2010) Computational identification of root hair-specific genes in Arabidopsis. Plant Signal Behav 5:1407–1418
Dang ZH, Zheng LL, Wang J, Gao Z, Wu SB, Qi Z, Wang YC (2013) Transcriptomic profiling of the salt-stress response in the wild recretohalophyte Reaumuria trigyna. BMC Genom 14:29
Ding F, Chen M, Sui N, Wang BS (2010a) Ca2+ significantly enhanced development and salt-secretion rate of salt glands of Limonium bicolor under NaCl treatment. S Afr J Bot 76:95–101
Ding F, Yang JC, Yuan F, Wang BS (2010b) Progress in mechanism of salt excretion in recretohalopytes. Front Biol 5:164–170
Dschida W, Platt-Aloia K, Thomson W (1992) Epidermal peels of Avicennia germinans (L.) Stearn: a useful system to study the function of salt glands. Ann Bot Lond 70:501–509
Edgar R, Domrachev M, Lash AE (2002) Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res 30:207–210
Faraday CD, Quinton PM, Thomson WW (1986) Ion fluxes across the transfusion zone of secreting Limonium salt glands determined from secretion rates, transfusion zone areas and plasmodesmatal frequencies. J Exp Bot 37:482–494
Fendrych M, Synek L, Pečenková T, Drdová EJ, Sekereš J, de Rycke R, Nowack MK, Žárský V (2013) Visualization of the exocyst complex dynamics at the plasma membrane of Arabidopsis thaliana. Mol Biol Cell 24:510–520
Flowers TJ, Colmer TD (2008) Salinity tolerance in halophytes. New Phytol 179:945–963
Flowers T, Troke P, Yeo A (1977) The mechanism of salt tolerance in halophytes. Annu Rev Plant Physiol 28:89–121
Gao Y, Gong X, Cao W, Zhao J, Fu L, Wang X, Schumaker KS, Guo Y (2008) SAD2 in Arabidopsis functions in trichome initiation through mediating GL3 function and regulating GL1, TTG1 and GL2 expression. J Integr Plant Biol 50:906–917
Hernandez JA, Jimenez A, Mullineaux P, Sevilia F (2000) Tolerance of pea (Pisum sativum L.) to long-term salt stress is associated with induction of antioxidant defences. Plant Cell Environ 23:853–862
Horie T, Hauser F, Schroeder JI (2009) HKT transporter-mediated salinity resistance mechanisms in Arabidopsis and monocot crop plants. Trends Plant Sci 14:660–668
Hu H, Dai M, Yao J, Xiao B, Li X, Zhang Q, Xiong L (2006) Overexpressing a NAM, ATAF, and CUC (NAC) transcription factor enhances drought resistance and salt tolerance in rice. Proc Natl Acad Sci USA 103:12987–12992
Huang J, Yang X, Wang MM, Tang H-J, Ding LY, Shen Y, Zhang HS (2007) A novel rice C2H2-type zinc finger protein lacking DLN-box/EAR-motif plays a role in salt tolerance. BBA Gene Struct Expr 1769:220–227
Ito M, Araki S, Matsunaga S, Itoh T, Nishihama R, Machida Y, Doonan JH, Watanabe A (2001) G2/M-phase-specific transcription during the plant cell cycle is mediated by c-Myb-like transcription factors. Plant Cell 13:1891–1905
Jiang C, Belfield EJ, Cao Y, Smith JA, Harberd NP (2013) An Arabidopsis soil-salinity-tolerance mutation confers ethylene-mediated enhancement of sodium/potassium homeostasis. Plant Cell 25:3535–3552
Karimi SH, Ungar IA (1989) Development of epidermal salt hairs in Atriplex triangularis Willd. in response to salinity, light intensity, and aeration. Bot Gaz 150:68–71
Klambauer G, Unterthiner T, Hochreiter S (2013) DEXUS: identifying differential expression in RNA-Seq studies with unknown conditions. Nucleic Acids Res 41:e198–e198
Kong L, Zhang Y, Ye ZQ, Liu XQ, Zhao SQ, Wei L, Gao G (2007) CPC: assess the protein-coding potential of transcripts using sequence features and support vector machine. Nucleic Acids Res 35:W345–W349
Kuwabara A, Nagata T (2006) Cellular basis of developmental plasticity observed in heterophyllous leaf formation of Ludwigia arcuata (Onagraceae). Planta 224:761–770
Lan WZ, Wang W, Wang SM, Li LG, Buchanan BB, Lin HX, Gao JP, Luan S (2010) A rice high-affinity potassium transporter (HKT) conceals a calcium-permeable cation channel. Proc Natl Acad Sci USA 107:7089–7094
Langmead B, Trapnell C, Pop M, Salzberg SL (2009) Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol 10(3):R25
Levering CA, Thomson WW (1971) The ultrastructure of the salt gland of Spartina foliosa. Planta 97:183–196
Li B, Dewey CN (2011) RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinform 12:323
Li F, Guo S, Zhao Y, Chen D, Chong K, Xu Y (2010) Overexpression of a homopeptide repeat-containing bHLH protein gene (OrbHLH001) from Dongxiang Wild Rice confers freezing and salt tolerance in transgenic Arabidopsis. Plant Cell Rep 29:977–986
Liphschitz N, Waisel Y (1982) Adaptation of plants to saline environments: salt excretion and glandular structure. In: Sen DN, Rajpurohit KS (eds) Tasks for Vegetation Science, vol 2. Dr Junk W Publ, The Hague, pp 197–214
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25:402–408
Lockhart J (2013) Salt of the earth: ethylene promotes salt tolerance by enhancing Na/K homeostasis. Plant Cell 25:3150
Luttge U (1971) Structure and function of plant glands. Ann Rev Plant Physiol 22:23–44
Meinke DW (1994) Seed development in Arabidopsis thaliana. In: Meyerowitz EM, Somerville CR (eds) Arabidopsis. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, pp 253–298
Meng L, Wong JH, Feldman LJ, Lemaux PG, Buchanan BB (2010) A membrane-associated thioredoxin required for plant growth moves from cell to cell, suggestive of a role in intercellular communication. Proc Natl Acad Sci USA 107:3900–3905
Munns R, Tester M (2008) Mechanisms of salinity tolerance. Annu Rev Plant Biol 59:651–681
Nadeau JA (2009) Stomatal development: new signals and fate determinants. Curr Opin Plant Biol 12:29–35
Naidoo Y, Naidoo G (1998) Salt glands of Sporobolus virginicus: morphology and ultrastructure. S Afr J Bot 64:198–204
Nass R, Cunningham KW, Rao R (1997) Intracellular sequestration of sodium by a novel Na+/H+ exchanger in yeast is enhanced by mutations in the plasma membrane H+-ATPase insights into mechanisms of sodium tolerance. J Biol Chem 272:26145–26152
Pandey S, Nelson DC, Assmann SM (2009) Two novel GPCR-type G proteins are abscisic acid receptors in Arabidopsis. Cell 136:136–148
Pang CH, Zhang SJ, Gong ZZ, Wang BS (2005) NaCl treatment markedly enhances H2O2-scavenging system in leaves of halophyte Suaeda salsa. Physiol Plant 125:490–499
Pardo JM, Cubero B, Leidi EO, Quintero FJ (2006) Alkali cation exchangers: roles in cellular homeostasis and stress tolerance. J Exp Bot 57:1181–1199
Payton S, Fray RG, Brown S, Grierson D (1996) Ethylene receptor expression is regulated during fruit ripening, flower senescence and abscission. Plant Mol Biol 31:1227–1231
Pillitteri LJ, Bogenschutz NL, Torii KU (2008) The bHLH protein, MUTE, controls differentiation of stomata and the hydathode pore in Arabidopsis. Plant Cell Physiol 49:934–943
Qi T, Song S, Ren Q, Wu D, Huang H, Chen Y, Fan M, Peng W, Ren C, Xie D (2011) The jasmonate-ZIM-domain proteins interact with the WD-repeat/bHLH/MYB complexes to regulate jasmonate-mediated anthocyanin accumulation and trichome initiation in Arabidopsis thaliana. Plant Cell 23:1795–1814
Rozema J, Flowers T (2008) Crops for a salinized world. Science 322:1478–1480
Sager R, Lee JY (2014) Plasmodesmata in integrated cell signalling: insights from development and environmental signals and stresses. J Exp Bot eru365:6337–6358
Schellmann S, Schnittger A, Kirik V, Wada T, Okada K, Beermann A, Thumfahrt J, Jürgens G, Hülskamp M (2002) TRIPTYCHON and CAPRICE mediate lateral inhibition during trichome and root hair patterning in Arabidopsis. EMBO J 21:5036–5046
Shi HZ, Ishitani M, Kim C, Zhu JK (2000) The Arabidopsis thaliana salt tolerance gene SOS1 encodes a putative Na+/H+ antiporter. Proc Natl Acad Sci USA 97:6896–6901
Shimony C, Fahn A (1968) Light and electron microscopical studies on the structure of salt glands of Tamarix aphylla L. J Linn Soc Lond Bot 60:283–288
Tan WK, Lin Q, Lim TM, Kumar P, Loh CS (2013) Dynamic secretion changes in the salt glands of the mangrove tree species Avicennia officinalis in response to a changing saline environment. Plant Cell Environ 36:1410–1422
Thomson WW, Platt-Aloia K (1985) The ultrastructure of the plasmodesmata of the salt glands of Tamarix as revealed by transmission and freeze-fracture electron microscopy. Protoplasma 125:13–23
Tsukagoshi H, Suzuki T, Nishikawa K, Agarie S, Ishiguro S, Higashiyama T (2015) RNA-seq analysis of the response of the halophyte, Mesembryanthemum crystallinum (ice plant) to high salinity. PLoS One 10(2):e0118339
Uno Y, Furihata T, Abe H, Yoshida R, Shinozaki K, Yamaguchi-Shinozaki K (2000) Arabidopsis basic leucine zipper transcription factors involved in an abscisic acid-dependent signal transduction pathway under drought and high-salinity conditions. Proc Natl Acad Sci USA 97:11632–11637
Van Camp W, Capiau K, Van Montagu M, Inze D, Slooten L (1996) Enhancement of oxidative stress tolerance in transgenic tobacco plants overproducing Fe-superoxide dismutase in chloroplasts. Plant Physiol 112:1703–1714
Van der Does D, Leon-Reyes A, Koornneef A, Van Verk MC, Rodenburg N, Pauwels L, Goossens A, Körbes AP, Memelink J, Ritsema T (2013) Salicylic acid suppresses jasmonic acid signaling downstream of SCFCOI1-JAZ by targeting GCC promoter motifs via transcription factor ORA59. Plant Cell 25:744–761
Verslues PE, Zhu JK (2007) New developments in abscisic acid perception and metabolism. Curr Opin Plant Biol 10:447–452
Waisel Y (1972) Biology of halophytes. Academic Press, New York, pp 184–193
Wang B, Lüttge U, Ratajczak R (2001) Effects of salt treatment and osmotic stress on V-ATPase and V-PPase in leaves of the halophyte Suaeda salsa. J Exp Bot 52:2355–2365
Wang YC, Ma H, Liu GF, Xu CX, Zhang DW, Ban QY (2008) Analysis of gene expression profile of Limonium bicolor under NaHCO3 stress using cDNA microarray. Plant Mol Biol Rep 26:241–254
Warnes G, Bolker B, Bonebakker L, Gentleman R, Huber W, Liaw A, Lumley T, Maechler M, Magnusson A, Moeller S (2010) gplots: Various R programming tools for plotting data. R package version 2.8.0
Xu ZY, Kim SY, Kim DH, Dong T, Park YM, Jin JB, Joo SH, Kim SK, Hong JC, Hwang D (2013) The Arabidopsis NAC transcription factor ANAC096 cooperates with bZIP-type transcription factors in dehydration and osmotic stress responses. Plant Cell 25:4708–4724
Yamamoto N, Takano T, Tanaka K, Ishige T, Terashima S, Endo C, Kurusu T, Yajima S, Yano K, Tada Y (2015) Comprehensive analysis of transcriptome response to salinity stress in the halophytic turf grass Sporobolus virginicus. Front Plant Sci 6:241
Yu H, Chen X, Hong YY, Wang Y, Xu P, Ke SD, Liu HY, Zhu JK, Oliver DJ, Xiang CB (2008) Activated expression of an Arabidopsis HD-START protein confers drought tolerance with improved root system and reduced stomatal density. Plant Cell 20:1134–1151
Yuan F, Chen M, Leng BY, Wang BS (2013) An efficient autofluorescence method for screening Limonium bicolor mutants for abnormal salt gland density and salt secretion. S Afr J Bot 88:110–117
Yuan F, Chen M, Yang JC, Leng BY, Wang BS (2014) A system for the transformation and regeneration of the recretohalophyte Limonium bicolor. In Vitro Cell Dev Plant 50:610–617
Yuan F, Lyv MJ, Leng BY, Zheng GY, Feng ZT, Li PH, Zhu XG, Wang BS (2015) Comparative transcriptome analysis of developmental stages of the Limonium bicolor leaf generates insights into salt gland differentiation. Plant Cell Environ 38:1637–1657
Zhu JK (2002) Salt and drought stress signal transduction in plants. Annu Rev Plant Biol 53:247–273
Zhu JK (2003) Regulation of ion homeostasis under salt stress. Curr Opin Plant Biol 6:441–445
Ziegler H, Lüttge U (1967) Die Salzdrüsen von Limonium vulgare. Planta 74:1–17
Acknowledgments
This work has been supported by the NSFC (National Natural Science Research Foundation of China, Project Nos. 31570251 and 31070158), Programs Foundation of Ministry of Education of China (20123704130001), Natural Science Research Foundation of Shandong Province (ZR2014CZ002), Young Scientist Research Award Fund of Shandong Province (BS2015SW027), and Technology Plan of Universities of Shandong Province (J15LE08).
Author contributions
B.W. and X.Z. designed the experiments. F.Y., M.A.L. and B.L. performed the experiments. F.Y. and M.A.L. analyzed the data. F.Y., M.A.L. and B.W. wrote the article. All authors read and approved the final manuscript.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors have declared that no competing interests exist.
Additional information
Fang Yuan and Ming-Ju Amy Lyu have contributed equally to this work.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Yuan, F., Lyu, MJ.A., Leng, BY. et al. The transcriptome of NaCl-treated Limonium bicolor leaves reveals the genes controlling salt secretion of salt gland. Plant Mol Biol 91, 241–256 (2016). https://doi.org/10.1007/s11103-016-0460-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11103-016-0460-0