Abstract
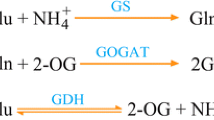
Glutamate decarboxylase (GAD, EC 4.1.1.15) has been suggested to be a key, regulatory point in the biosynthesis of γ-aminobutyrate (GABA) and in the utilization of citric acid through GABA shunt pathway. In this study we discovered two GAD genes, named as CsGAD1 and CsGAD2, in citrus genome database and then successfully cloned. Both CsGAD1 and CsGAD2 have a putative pyridoxal 5-phosphate binding domain in the middle region and a putative calmodulin-binding domain at the carboxyl terminus. Gene structure analysis showed that much difference exists in the size of exons and introns or in cis-regulatory elements in promoter region between the two GAD genes. Gene expression indicated that CsGAD1 transcript was predominantly expressed in flower and CsGAD2 transcript was predominantly expressed in fruit juice sacs; in the ripening fruit, CsGAD1 transcript level was at least 2-time higher than CsGAD2 transcript level. Moreover, CsGAD1 transcript level was increased significantly along with the increase of GAD activity and accompanied by a significant decrease of titratable acid (TA), suggesting that it is CsGAD1 rather than CsGAD2 plays a role in the citric acid utilization during fruit ripening. In addition, injection of abscisic acid and foliar spray of K2SO4 significantly increased the TA content of Satsuma mandarin, and significantly decreased GAD activity as well as CsGAD1 transcript, further suggesting the important role of CsGAD1 in the citrate utilization of citrus fruit.






Similar content being viewed by others
References
Marenco S, Savostyanova AA, van der Veen JW, Geramita M, Stern A, Barnett AS, Kolachana B, Radulescu E, Zhang F, Callicott JH, Straub RE, Shen J, Weinberger DR (2010) Genetic modulation of GABA levels in the anterior cingulate cortex by GAD1 and COMT. Neuropsychopharmacology 35(8):1708–1717
Shimajiri Y, Oonishi T, Ozaki K, Kainou K, Akama K (2013) Genetic manipulation of the γ-aminobutyric acid (GABA) shunt in rice: overexpression of truncated glutamate decarboxylase (GAD2) and knockdown of γ-aminobutyric acid transaminase (GABA-T) lead to sustained and high levels of GABA accumulation in rice kernels. Plant Biotechnol J 11(5):594–604
Akihiro T, Koike S, Tani R, Tominaga T, Watanabe S, Iijima Y, Aoki K, Shibata D, Ashihara H, Matsukura C, Akama K, Fujimura T, Ezura H (2008) Biochemical mechanism on GABA accumulation during fruit development in tomato. Plant Cell Physiol 49(9):1378–1389
Cercós M, Soler G, Iglesias D, Gadea J, Forment J, Talón M (2006) Global analysis of gene expression during development and ripening of citrus fruit flesh. A proposed mechanism for citric acid utilization. Plant Mol Biol 62(4–5):513–527
Degu A, Hatew B, Nunes-Nesi A, Shlizerman L, Zur N, Katz E, Fernie AR, Blumwald E, Sadka A (2011) Inhibition of aconitase in citrus fruit callus results in a metabolic shift towards amino acid biosynthesis. Planta 234(3):501–513
Shelp BJ, Bown AW, McLean MD (1999) Metabolism and functions of gamma-aminobutyric acid. Trends Plant Sci 4(11):446–452
Fait A, Fromm H, Walter D, Galili G, Fernie AR (2008) Highway or byway: the metabolic role of the GABA shunt in plants. Trends Plant Sci 13(1):14–19
Bouché N, Fromm H (2004) GABA in plants: just a metabolite? Trends Plant Sci 9(3):110–115
Shelp BJ, Bozzo GG, Zarei A, Simpson JP, Trobacher CP, Allan WL (2012) Strategies and tools for studying the metabolism and function of gamma-aminobutyrate in plants: II integrated analysis. Botany 90:781–793
Adeghate E, Ponery AS (2002) GABA in the endocrine pancreas: cellular localization and function in normal and diabetic rats. Tissue Cell 34(1):1–6
Hayakawa K, Kimura M, Kasaha K, Matsumoto K, Sansawa H, Yamori Y (2004) Effect of a gamma-aminobutyric acid-enriched dairy product on the blood pressure of spontaneously hypertensive and normotensive Wistar–Kyoto rats. Br J Nutr 92(3):411–417
Amin Z, Mason GF, Cavus I, Krystal JH, Rothman DL, Epperson CN (2006) The interaction of neuroactive steroids and GABA in the development of neuropsychiatric disorders in women. Pharmacol Biochem Behav 84(4):635–643
Akama K, Kanetou J, Shimosaki S, Kawakami K, Tsuchikura S, Takaiwa F (2009) Seed-specific expression of truncated OsGAD2 produces GABA-enriched rice grains that influence a decrease in blood pressure in spontaneously hypertensive rats. Transgenic Res 18(6):865–876
Gladkevich A, Korf J, Hakobyan VP, Melkonyan KV (2006) The peripheral GABAergic system as a target in endocrine disorders. Auton Neurosci 124(1–2):1–8
Ueno H (2000) Enzymatic and structural aspects on glutamate decarboxylase. J Mol Catal B Enzym 10:67–79
Snedden WA, Koustia N, Baum G, Fromm H (1996) Activation of a recombinant petunia glutamate decarboxylase by calcium/calmodulin or by a monoclonal antibody which recognizes the calmodulin binding domain. J Biol Chem 271:4148–4153
Gut H, Dominici P, Pilati S, Astegno A, Petoukhov MV, Svergun DI, Gr uuml tter MG, Capitani G (2009) A common structural basis for pH- and calmodulin-mediated regulation in plant glutamate decarboxylase. J Mol Biol 392:334–351
Akama K, Takaiwa F (2007) C-terminal extension of rice glutamate decarboxylase (OsGAD2) functions as an autoinhibitory domain and overexpression of a truncated mutant results in the accumulation of extremely high levels of GABA in plant cells. J Exp Bot 58:2699–2707
Zik M, Arazi T, Snedden WA, Fromm H (1998) Two isoforms of glutamate decarboxylase in Arabidopsis are regulated by calcium/calmodulin and differ in organ distribution. Plant Mol Biol 37:967–975
Baum G, Chen Y, Arazi T, Takatsuji H, Fromm H (1993) A plant glutamate decarboxylase containing a calmodulin binding domain. Cloning, sequence, and functional analysis. J Biol Chem 268(26):19610–19617
Gallego P, Whotton L, Picton S, Grierson D, Gray J (1995) A role for glutamate decarboxylase during tomato ripening: the characterisation of a cDNA encoding a putative glutamate decarboxylase with a calmodulin-binding site. Plant Mol Biol 27(6):1143–1151
Johnson BS, Singh NK, Cherry JH, Locy RD (1997) Purification and characterization of glutamate decarboxylase from cowpea. Phytochemistry 46(1):39–44
Bouché N, Fait A, Zik M, Fromm H (2004) The root-specific glutamate decarboxylase (GAD1) is essential for sustaining GABA levels in Arabidopsis. Plant Mol Biol 55(3):315–325
Turano FJ, Fang TK (1998) Characterization of two glutamate decarboxylase cDNA clones from Arabidopsis. Plant Physiol 117(4):1411–1421
Yevtushenko DP, McLean MD, Peiris S, Van Cauwenberghe OR, Shelp BJ (2003) Calcium/calmodulin activation of two divergent glutamate decarboxylases from tobacco. J Exp Bot 54:2001–2002
Oh S-H, Choi W-G, Lee I-T, Yun SJ (2005) Cloning and characterization of a rice cDNA encoding glutamate decarboxylase. J Biochem Mol Biol 38(5):595–601
Zhuang Y, Ren G, He C, Li X, Meng Q, Zhu C, Wang R, Zhang J (2010) Cloning and characterization of a maize cDNA encoding gutamate decarboxylase. Plant Mol Biol Rep 28(4):620–626
Molina-Rueda JJ, Pascual MB, Cánovas, Francisco M, Gallardo F (2010) Characterization and developmental expression of a glutamate decarboxylase from maritime pine. Planta 232(6):1471–1483
Trobacher C, Zarei A, Liu J, Clark S, Bozzo G, Shelp B (2013) Calmodulin-dependent and calmodulin-independent glutamate decarboxylases in apple fruit. BMC Plant Biol 13(1):144
Arazi T, Baum G, Snedden WA, Shelp BJ, Fromm H (1995) Molecular and biochemical analysis of calmodulin interactions with the calmodulin-binding domain of plant glutamate decarboxylase. Plant Physiol 108:551–561
McLean M, Yevtushenko D, Deschene A, Van Cauwenberghe O, Makhmoudova A, Potter J, Bown A, Shelp B (2003) Overexpression of glutamate decarboxylase in transgenic tobacco plants confers resistance to the northern root-knot nematode. Mol Breed 11(4):277–285
Lee J-H, Kim Y-J, Jeong D-Y, Sathiyaraj G, Pulla R, Shim J-S, In J-G, Yang D-C (2010) Isolation and characterization of a Glutamate decarboxylase (GAD) gene and their differential expression in response to abiotic stresses from Panax ginseng C. A. Meyer. Mol Biol Rep 37(7):3455–3463
Baum G, Lev-Yadun S, Fridmann Y, Arazi T, Katsnelson H, Zik M, Fromm H (1996) Calmodulin binding to glutamate decarboxylase is required for regulation of glutamate and GABA metabolism and normal development in plants. EMBO J 15:2988–2996
Chen Y, Baum G, Fromm H (1994) The 58-kilodalton calmodulin-binding glutamate decarboxylase is a ubiquitous protein in petunia organs and its expression is developmentally regulated. Plant Physiol 106(4):1381–1387
Baldwin EA (1993) Citrus fruit. In: Seymour GB, Taylor JE, Tucker GA (eds) Biochemistry of fruit ripening. Chapman & Hall, London, pp 107–149
Chen M, Jiang Q, Yin X-R, Lin Q, Chen J-Y, Allan AC, Xu C-J, Chen K-S (2012) Effect of hot air treatment on organic acid- and sugar-metabolism in Ponkan (Citrus reticulata) fruit. Sci Horti 147:118–125
Zhou GF, Peng SA, Liu YZ, Wei QJ, Han J, Islam MZ (2014) The physiological and nutritional responses of seven different citrus rootstock seedlings to boron deficiency. Trees 28(1):295–307
Kojima K, Yamada Y, Yamamoto M (1995) Effects of abscisic acid injection on sugar and organic acid contents of citrus fruit. J Jpn Soc Hortic Sci 64:17–21
Yang S-Y, Lu Z-X, Lu F-X, Bie X-M, Sun L-J (2006) Colorimetric determination of GABA in GAD activity assay. Food Sci 7:205–209
Xu Q, Chen LL, Ruan X, Chen D, Zhu A, Chen C, Bertrand D, Jiao WB, Hao BH, Lyon MP, Chen J, Gao S, Xing F, Lan H, Chang JW, Ge X, Lei Y, Hu Q, Miao Y, Wang L, Xiao S, Biswas MK, Zeng W, Guo F, Cao H, Yang X, Xu XW, Cheng YJ, Xu J, Liu JH, Luo OJ, Tang Z, Guo WW, Kuang H, Zhang HY, Roose ML, Nagarajan N, Deng XX, Ruan Y (2012) The draft genome of sweet orange (Citrus sinensis). Nat Genet 45(1):59–66
Liu YZ, Liu Q, Tao NG, Deng XX (2006) Efficient isolation of RNA from fruit peel and pulp of ripening navel orange (Citrus sinensis Osbeck). J Huazhong Agric Univ 25:300–304
Koressaar T, Remm M (2007) Enhancements and modifications of primer design program Primer3. Bioinformatics 23(10):1289–1291
Guo A-Y, Zhu Q-H, Chen X, Luo J-C (2007) GSDS: a gene structure display server. Yi Chuan 29(8):1023
Higo K, Ugawa Y, Iwamoto M, Korenaga T (1999) Plant cis-acting regulatory DNA elements (PLACE) database: 1999. Nucleic Acids Res 27(1):297–300
Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S (2011) MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28(10):2731–2739
Livak KJ, Schmittigen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(-delta delta C(T)) method. Methods 25:402–408
Kempa S, Krasensky J, Dal Santo S, Kopka J, Jonak C (2008) A central role of abscisic acid in stress-regulated carbohydrate metabolism. PLoS One 3(12):e3935
Navarro JM, Pérez-Pérez JG, Romero P, Botía P (2010) Analysis of the changes in quality in mandarin fruit, produced by deficit irrigation treatments. Food Chem 119(4):1591–1596
Hockema BR, Etxeberria E (2001) Metabolic contributors to drought-enhanced accumulation of sugars and acids in oranges. J Am Soc Hortic Sci 126(5):599–605
Jiang N, Jin L-F, Teixeira da Silva JA, Islam MDZ, Gao H-W, Liu Y-Z, Peng S-A (2014) Activities of enzymes directly related with sucrose and citric acid metabolism in citrus fruit in response to soil plastic film mulch. Sci Hortic 168:73–80
Bastías A, López-Climent M, Valcárcel M, Rosello S, Gómez-Cadenas A, Casaretto JA (2011) Modulation of organic acids and sugar content in tomato fruits by an abscisic acid-regulated transcription factor. Physiol Plant 141(3):215–226
Leonhardt N, Kwak JM, Robert N, Waner D, Leonhardt G, Schroeder JI (2004) Microarray expression analyses of Arabidopsis guard cells and isolation of a recessive abscisic acid hypersensitive protein phosphatase 2C mutant. Plant Cell Online 16(3):596–615
Li Y, Lee KK, Walsh S, Smith C, Hadingham S, Sorefan K, Cawley G, Bevan MW (2006) Establishing glucose-and ABA-regulated transcription networks in Arabidopsis by microarray analysis and promoter classification using a Relevance Vector Machine. Genome Res 16(3):414–427
Alva AK, Paramasivam S, Dou H, Sajwan KS (2006) Potassium management for optimizing citrus production and quality. Int J Fruit Sci 6:3–43
Tsay Y-F, Ho C-H, Chen H-Y, Lin S-H (2011) Integration of nitrogen and potassium signaling. Annu Rev Plant Biol 62:207–226
Liu H, Shi C, Zhang H, Wang Z, Chai S (2013) Effects of potassium on yield, photosynthate distribution, enzymes’ activity and aba content in storage roots of sweet potato (‘Ipomoea batatas’ Lam.). Aust J Crop Sci 7(6):735–743
Haeder HE, Beringer H (1981) Influence of potassium nutrition and water stress on the content of abscisic acid in grains and flag leaves of wheat during grain development. J Sci Food Agric 32(6):552–556
Marrush M, Yamaguchi M, Saltveit ME (1998) Effect of potassium nutrition during bell pepper seed development on vivipary and endogenous levels of abscisic acid (ABA). J Am Soc Hortic Sci 123(5):925–930
Acknowledgments
This work was supported by Hubei Province Natural Science Foundation (No. ZRY0116) and the National Natural Science Foundation of China (No. 31372012).
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Liu, X., Hu, XM., Jin, LF. et al. Identification and transcript analysis of two glutamate decarboxylase genes, CsGAD1 and CsGAD2, reveal the strong relationship between CsGAD1 and citrate utilization in citrus fruit. Mol Biol Rep 41, 6253–6262 (2014). https://doi.org/10.1007/s11033-014-3506-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11033-014-3506-x