Abstract
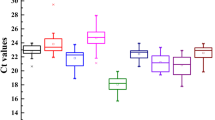
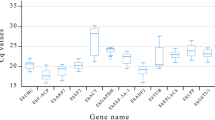
Accurate quantification of transcript profiling with quantitative real time polymerase chain reaction (qRT-PCR) relies on the reliable normalization of an appropriate reference gene. This study reported the identification and validation of nine reference genes, including β-tubulin (β-TUB), elongation factor 1 alpha (EF-1α), elongation factor 1 beta (EF-1β), glyceraldehyde-3-phosphate dehydrogenase (GAPDH), ubiquitin (UBQ), actin 1/2(ACT-1 and ACT-2), 18S rRNA, and 26S rRNA, from Anoectochilus roxburghii (Wall.) Lindl., a valuable herb remedy widely used for various diseases treatment in traditional Chinese medicine. Transcriptional levels of the candidate reference genes were examined using qRT-PCR analysis and revealed differential expression of the genes in the leaf, stem, root, flower, and peduncle tissues. The relative quantities data were subjected to geNorm software for ranking the expression stability of the reference genes and the results showed that EF-1β and ACT-2 were the two best stable genes whereas GAPDH and 26S rRNA did not favor normalization of qRT-PCR in these tissues. The expression pattern of a squalene synthase encoding gene (SS) was also determined in parallel. The analyses were in great consistency when the qRT-PCR data was normalized to the expression of each or both of EF-1β and ACT-2 as the internal control, further confirming the reliability of EF-1β and ACT-2 as the best internal control. The present study provided the first important clues for accurate data normalization in transcript profiling in A. roxburghii, which will be essential to further functional genomics study in the valuable medicinal plant.



Similar content being viewed by others
Abbreviations
- ACT-1:
-
Actin 1
- ACT-2:
-
Actin 2
- CT:
-
Threshold value
- E :
-
PCR efficiency
- EF-1α :
-
Elongation factor 1 alpha
- EF-1β :
-
Elongation factor 1 beta
- GAPDH:
-
Glyceraldehyde-3-phosphate dehydrogenase
- Q :
-
Relative quantities
- qRT-PCR:
-
Quantitative reverse transcription PCR
- RT-PCR:
-
Reverse transcription polymerase chain reaction
- sqPCR:
-
Semi-quantitative PCR
- UBQ:
-
Ubiquitin
- 18S rRNA:
-
18S ribosomal RNA
- 26S rRNA:
-
26S ribosomal RNA
- β-TUB:
-
β-Tubulin
References
Gutierrez L (2008) Towards a systematic validation of references in real-time RT-PCR. Plant Cell 20:1734–1735
Bustin SA (2002) Quantification of mRNA using real-time reverse transcription PCR (RT-PCR): trends and problems. J Mol Endocrinol 29:23–39
Hacquard S, Veneault-Fourrey C, Delaruelle C, Frey P, Martin F, Duplessis S (2011) Validation of Melampsora larici-populina reference genes for in planta RT-quantitative PCR expression profiling during time-course infection of poplar leaves. Physiol Mol Plant Pathol 75:106–112
Gachon C, Mingam A, Charrier B (2004) Real-time PCR: what relevance to plant studies? J Exp Bot 55:1445–1454
Czechowski T, Stitt M, Altmann T, Udvardi MK, Scheible W (2005) Genome-wide identification and testing of superior reference genes for transcript normalization in Arabidopsis. Plant Physiol 139:5–17
Løvdal T, Lillo C (2009) Reference gene selection for quantitative real-time PCR normalization in tomato subjected to nitrogen, cold, and light stress. Anal Biochem 387:238–242
Guénin S, Mauriat M, Pelloux J, Wuytswinkel OV, Bellini C, Gutierrez L (2009) Normalization of qRT-PCR data: the necessity of adopting a systematic, experimental conditions specific, validation of references. J Exp Bot 60:487–493
Vandesompele J, De Preter K, Pattyn F, Poppe B, Van Roy N, De Paepe A, Speleman F (2002) Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol 3(7):0034.1–0034.11
Andersen CL, Jensen JL, Ørntoft TF (2004) Normalization of real-time quantitative reverse transcription-PCR data: a model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Res 64:5245–5250
Pfaffl MW, Tichopád A, Prgomet C (2004) Determination of stable housekeeping genes, differentially regulated target genes and sample integrity: BestKeeper–Excel-based using pair-wise correlations. Biotechnol Lett 26:509–515
Mallona I, Lischewski S, Weiss J (2010) Validation of reference genes for quantitative real-time PCR during leaf and flower development in Petunia hybrida. BMC Plant Biol 10:4
Mahoney DJ, Carey K, Fu MH, Snow R, Cameron-Smith D, Parise G, Tarnopolsky MA (2004) Real-time RT-PCR analysis of housekeeping genes in human skeletal muscle following acute exercise. Physiol Genomics 18:226–231
Zhong Q, Zhang Q, Wang Z, Qi J, Chen Y, Li S, Sun Y, Li C, Lan X (2008) Expression profiling and validation of potential reference genes during Paralichthys olivaceus embryogenesis. Mar Biotechnol 10:310–318
Yuwen Y, Dong Z, Wang Q, Sun X, Shi C, Chen G (2011) Evaluation of endogenous reference genes for analysis of gene expression with real-time RT-PCR during planarian regeneration. Mol Biol Rep 38(7):16–21
Jain M, Nijhawan A, Tyagi AK, Khurana JP (2006) Validation of housekeeping genes as internal control for studying gene expression in rice by quantitative real-time PCR. Biochem Biophys Res Commun 345:646–651
Jian B, Liu B, Bi Y, Hou W, Wu C, Han T (2008) Validation of internal control for gene expression study in soybean by quantitative real-time PCR. BMC Mol Biol 9:59
Cruz F, Kalaoun S, Nobile P, Colombo C, Almeida J, Barros LMG, Romano E, Grossi-de-Sá MF, Vaslin M, Alves-Ferreira M (2009) Evaluation of coffee reference genes for relative expression studies by quantitative real-time RT-PCR. Mol Breeding 23(4):607–616
Zhang Y, Cai J, Ruan H, Pi H, Wu J (2007) Antihyperglycemic activity of kinsenoside, a high yielding constituent from Anoectochilus roxburghii in streptozotocin diabetic rats. J Ethnopharmacol 14(2):141–145
He C, Wang C, Guo S, Yang J, Xiao P (2006) A novel flavonoid glucoside from Anoectochilus roxberghii (Wall.) Lindl. J Integr Plant Biol 48(3):359–363
Surh YJ, Chus KS, Cha HH, Han SS, Keum YS, Park KK, Lee SS (2001) Molecular mechanisms underling chemopreventive activities of anti-inflammatory phytochemicals: down-regulation of COX-2 and iNOS through suppression of NF-κB activation. Mutat Res 480–481:243–268
Du XM, Irino N, Furusho N, Hayashi J, Shoyama Y (2008) Pharmacologically active compounds in the Anoectochilus and Goodyera species. J Nat Med 62:132–148
Ikeuchi M, Yamaguchi K, Nishimura T, Yazawa K (2005) Effects of Anoectochilus formosanus on endurance capacity in mice. J Nutr Sci Vitaminol 51:40–44
Hsieh WT, Tsai CT, Wu JB, Hsiao HB, Yang LC, Lin WC (2011) Kinsenoside, a high yielding constituent from Anoectochilus formosanus, inhibits carbon tetrachloride induced Kupffer cells mediated liver damage. J Ethnopharmacol 135(2):440–449
Liao P, Zhou W, Zhang L, Wang J, Yan X, Zhang Y, Zhang R, Zhou G, Kai G (2009) Molecular cloning, characterization and expression analysis of a new gene encoding 3-hydroxy-3-methylglutaryl coenzyme A reductase from Salvia miltiorrhiza. Acta Physiol Plant 31(3):565–572
Huang JZ, Cheng TC, Wen PJ, Hsieh MH, Chen FC (2009) Molecular characterization of the Oncidium orchid HDR gene encoding 1-hydroxy-2-methyl-2-(E)-butenyl 4-diphosphate reductase, the last step of the methylerythritol phosphate pathway. Plant Cell Rep 28(10):1475–1486
Skipper M, Pedersen KB, Johansen LB, Frederiksen S, Irish VF, Johansen BB (2005) Identification and quantification of expression levels of three FRUITFULL-like MADS-box genes from the orchid Dendrobium thyrsiflorum (Reichb. f.). Plant Sci 169:579–586
Jia XY, Xu CY, Jing RL, Li RZ, Mao XG, Wang JP, Chang XP (2008) Molecular cloning and characterization of wheat calreticulin (CRT) gene involved in drought-stressed responses. J Exp Bot 59(4):739–751
Yang Y, Wu J, Zhu K, Liu L, Chen F, Yu D (2009) Identification and characterization of two chrysanthemum (Dendronthema × moriforlium) DREB genes, belonging to the AP2/EREBP family. Mol Biol Rep 36:71–81
Lu Q, Yang Q (2006) cDNA cloning and expression of anthocyanin biosynthetic genes in wild potato (Solanum pinnatisectum). Afr J Biotechnol 5(10):811–818
Li SH, Kuoh CS, Chen YH, Chen HH, Chen WH (2005) Osmotic sucrose enhancement of single-cell embryogenesis and transformation efficiency in Oncidium. Plant Cell Tiss Org 81:183–192
Rozen S, Skaletsky H (2000) Primer3 on the WWW for general users and for biologist programmers. Methods Mol Bio 132:365–386
Pfaffl MW (2001) A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res 29(9):e45
Yang Y, Hou S, Cui G, Chen S, Wei J, Huang L (2010) Characterization of reference genes for quantitative real-time PCR analysis in various tissues of Salvia miltiorrhiza. Mol Biol Rep 37:507–513
Dong L, Sui C, Liu Y, Yang Y, Wei J, Yang Y (2011) Validation and application of reference genes for quantitative gene expression analyses in various tissues of Bupleurum chinense. Mol Biol Rep 38(8):5017–5023
Yan J, Yuan F, Long G, Qin L, Deng Z (2011) Selection of reference genes for quantitative real-time RT-PCR analysis in citrus. Mol Biol Rep doi:10.1007/s11033-011-0925-9
Thellin O, ElMoualij B, Heinen E, Zorzi W (2009) A decade of improvements in quantification of gene expression and internal standard selection. Biotechnol Adv 27(4):323–333
Singh K, Raizada J, Bhardwaj P, Ghawana S, Rani A, Singh H, Kaul K, Kumar S (2004) 26S rRNA-based internal control gene primer pair for reverse transcription-polymerase chain reaction-based quantitative expression studies in diverse plant species. Anal Biochem 335:330–333
Lee MH, Jeong JH, Seo JW, Shin CG, Kim YS, In JG, Yang DC, Yi JS, Choi YE (2004) Enhanced triterpene and phytosterol biosynthesis in Panax ginseng overexpressing squalene synthase gene. Plant Cell Physiol 45(8):976–984
Kim TD, Han JY, Huh GH, Choi YE (2011) Expression and functional characterization of three squalene synthase genes associated with saponin biosynthesis in Panax ginseng. Plant Cell Physiol 52(1):125–137
Zhang L, Jing F, Li F, Li M, Wang Y, Wang G, Sun X, Tang K (2009) Development of transgenic Artemisia annua (Chinese wormwood) plants with an enhanced content of artemisinin, an effective anti-malarial drug, by hairpin-RNA-mediated gene silencing. Biotechnol Appl Biochem 52:199–207
Acknowledgments
This research was financially supported by the National Natural Sciences Foundation of China (No. 31101608 and 31070300), and the China Postdoctoral Science Foundation (No. 20100470244).
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Zhang, G., Zhao, M., Song, C. et al. Characterization of reference genes for quantitative real-time PCR analysis in various tissues of Anoectochilus roxburghii . Mol Biol Rep 39, 5905–5912 (2012). https://doi.org/10.1007/s11033-011-1402-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11033-011-1402-1