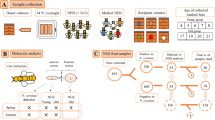
Abstract
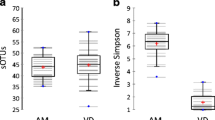
Tropilaelaps mercedesae is an ectoparasite of Apis mellifera in Asia and is considered a major threat to honey bee health. Herein, we used the Illumina MiSeq platform 16S rDNA Amplicon Sequencing targeting the V3–V4 regions and analysed the effects on the midgut bacterial communities of honey bees infested with T. mercedesae. The overall bacterial community in honey bees infested with T. mercedesae were observed at different developmental stages. Honey bee core intestinal bacterial genera such as Gilliamella, Lactobacillus and Frischella were detected. Tropilaelapsmercedesae infestation changed the bacterial communities in the midgut of A. mellifera. Tropilaelapsmercedesae-infested pupae had greatly increased relative abundances of Micrococcus and Sphingomonas, whereas T. mercedesae-infested 15-day-old workers had significantly reduced relative abundance of non-core microbes: Corynebacterium, Sphingomonas, Acinetobacter and Enhydrobacter compared to T. mercedesae-infested newly emerged bees. The bacterial community was significantly changed at the various T. mercedesae-infested developmental stages of A. mellifera. Tropilaelapsmercedesae infestation also changed the non-core bacterial community from larvae to newly emerged honey bees. Bacterial communities were significantly different between T. mercedesa-infested and non-mite-infested 15-day-old workers. Lactobacillus was dominant in T. mercedesae-infested 15-day-old workers compared to non-mite-infested 15-day-old workers.








Similar content being viewed by others
References
Anderson DL, Morgan MJ (2007) Genetic and morphological variation of bee parasitic Tropilaelaps mites (Acari: Laelapidae): new and re-defined species. Exp Appl Acarol 43:1–24
Anderson KE, Sheehan TH, Eckholm BJ, Mott BM, DeGrandi-Hoffman G (2011) An emerging paradigm of colony health: microbial balance of the honey bee and hive (Apis mellifera). Insect Soc 58:431–444
Anderson KE, Sheehan TH, Mott BM, Mott BM, Maes P, Snyder L, Schwan MR, Walton A, Jones BM, Corby-Harr V (2013) Microbial ecology of the hive and pollination landscape: bacterial associates from floral nectar, the alimentary tract and stored food of honey bees (Apis mellifera). PLoS ONE 8:e83125
Butler È, Alsterfjord M, Olofsson TC, Karlsson C, Malmström J, Olofsson AV (2013) Proteins of novel lactic acid bacteria from Apis mellifera mellifera: an insight into the production of known extra-cellular proteins during microbial stress. BMC Microbiol 13:235
Camphor ESW, Hashmi AA, Ritter W, Bowen ID (2005) Seasonal changes in mite (Tropilaelaps clareae) and honeybee (Apis mellifera) populations in apistan treated and untreated colonies. Apiacta 40:34–44
Chandler D, Sunderland KD, Ball BV, Davidson G (2001) Prospective biological control agents of Varroa destructor n. sp. an important pest of the European honeybee, Apis mellifera. Biocontrol Sci Technol 11:429–448
Crotti E, Sansonno L, Prosdocimi EM, Vacchini V, Hamdi C, Cherif A, Gonella E, Marzorati M, Balloi A (2013) Microbial symbionts of honeybees: a promising tool to improve honeybee health. N Biotechnol 30:716–722
Dainat B, Ken T, Berthoud H, Neumann P (2009) The ectoparasitic mite Tropilaelaps mercedesae (Acari, Laelapidae) as a vector of honeybee viruses. Insect Soc 56:40–43
de Guzman LI, Williams GR, Khongphinitbunjong K, Chantawannakul P (2017) Ecology, life history, and management of Tropilaelaps mites. J Econ Entomol 110:319
Disayathanoowat T, Young JP, Helgason T, Chantawannakul P (2012) T-RFLP analysis of bacterial communities in the midguts of Apis mellifera and Apis cerana honey bees in Thailand. FEMS Microbiol Ecol 79(2):273–281
Edgar RC (2004) MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32:1792–1797
Edgar RC, Haas BJ, Clemente JC, Quince C, Knight R (2011) UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 27:2194–2200
Ellegaard KM, Engel P (2015) Beyond 16S rRNA community profiling: intraspecies diversity in the gut microbiota. Front Microbiol 7:1475
Ellegaard KM, Tamarit D, Javelind E, Olofsson TC, Andersson SGE, Vásquez A (2016) Extensive intra-phylotype diversity in lactobacilli and bifidobacteria from the honeybee gut. BMC Genom 16:284
Engel P, Moran NA (2013) The gut microbiota of insects—diversity in structure and function. FEMS Microbiol Rev 37:699–735
Evans JD, Schwarz RS (2011) Bees brought to their knees: microbes affecting honey bee health. Trends Microbiol 19:614–620
Fei DL, Zhang HC, Diao QY, Jiang LL, Wang Q, Zhong Y, Fan ZB, Ma MX (2015) Codon optimization, expression in Escherichia coli, and immunogenicity of recombinant Chinese Sacbrood Virus (CSBV) structural proteins VP1, VP2, and VP3. PLoS ONE 10:e0128486
Gallai N, Salles JM, Settele J, Vaissière BE (2009) Economic valuation of the vulnerability of world agriculture confronted with pollinator decline. Ecol Econ 68:810–821
Gerth M, Hurst GDD (2017) Short reads from honey bee (Apis sp.) sequencing projects reflect microbial associate diversity. Peer J 5:e3529
Guo J, Jie W, Chen YP, Evans JD, Dai RG, Luo WH, Li JL (2015) Characterization of gut bacteria at different developmental stages of Asian honey bees, Apis cerana. J Invertebr Pathol 127:110–114
Haas BJ, Dirk G, Ashlee ME, Mike F, Doyle VW, Georgia G, Dawn C, Diana T, Sarah KH, Erica S, Barbara M, Todd ZD, Joseph FP, Rob K, Bruce WB (2011) Chimeric 16S rRNA sequence formation and detection in Sanger and 454-pyrosequenced PCR amplicons. Genome Res 21:494–504
Hroncova Z, Havlik J, Killer J, Doskocil I, Tyl J, Kamler M, Titera D, Hakl J, Mrazek J, Bunesova V, Rada V (2015) Variation in honey bee gut microbial diversity affected by ontogenetic stage, age and geographic location. PLoS ONE 10:e0118707
Hubert J, Erban T, Kamler M, Kopecky J, Nesvorna M, Hejdankova S, Titera D, Tyl J, Zurek L (2015) Bacteria detected in the honeybee parasitic mite Varroa destructor collected from beehive winter debris. J Appl Microbiol 119:640–654
Jia HR, Geng LL, Li YH, Wang Q, Diao QY, Zhou T, Dai PL (2016) The effects of Bt Cry1Ie toxin on bacterial diversity in the midgut of Apis mellifera ligustica (Hymenoptera: Apidae). Sci Rep 6:24664
Jia HR, Dai PL, Geng LL, Jack CJ, Li YH, Wu YY, Diao QY, Ellis JD (2017) No effect of Bt Cry1Ie toxin on bacterial diversity in the midgut of the Chinese honey bees, Apis cerana cerana (Hymenoptera, Apidae). Sci Rep 7:41688
Kanga LHB, Jones WA, James RR (2003) Field trials using the fungal pathogen, Metarhizium anisopliae (Deuteromycetes: Hyphomycetes) to control the ectoparasitic mite, Varroa destructor (Acari: Varroidae) in Honey Bee, Apis mellifera (Hymenoptera: Apidae) colonies. J Econ Entomol 96:1091–1099
Kanga LHB, Jones WA, Gracia C (2006) Efficacy of strips coated with Metarhizium anisopliae for control of Varroa destructor (Acari: Varroidae) in honey bee colonies in Texas and Florida. Exp Appl Acarol 40:249–258
Kapheim KM, Rao VD, Yeoman CJ, Wilson BA, White BA, Goldenfeld N, Robinson GE (2015) Caste-Specific differences in hindgut microbial communities of honey bees (Apis mellifera). PLoS ONE 10:e0123911
Kešnerová L, Moritz R, Engel P (2015) Bartonella apis sp. nov. a honey bee gut symbiont of the class Alphaproteobacteria. Int J Syst Evol Micr 66:414–421
Khongphinitbunjong K, Guzman LID, Burgett MD, Rinderer TE, Chantawannakul P (2012) Behavioral responses underpinning resistance and susceptibility of honeybees to Tropilaelaps mercedesae. Apidologie 43:590–599
Khongphinitbunjong K, Neumann P, Chantawannakul P, Williams GR (2016) The ectoparasitic mite Tropilaelaps mercedesae reduces western honey bee, Apis mellifera, longevity and emergence weight, and promotes deformed wing virus infections. Invertebr Pathol 137:38–42
Killer J, DubnáS Sedláček I, Švec P (2014) Lactobacillus apis sp. nov., from the stomach of honeybees (Apis mellifera), having an in vitro inhibitory effect on the causative agents of American and European foulbrood. Int J Syst Evol Microbiol 64:152–157
Koch H, Schmid-Hempel P (2011) Bacterial communities in central European bumblebees: low diversity and high specificity. Microbial Ecol 62:121–133
Kumar R, Kumar NR, Bhalla OP (1993) Studies on the development biology of Tropilaelaps clareae Delfinado and Baker (Acarina: Laelapidae) vis a vis the threshold stage in the life cycle of Apis mellifera Linn. (Hymenoptera: Apidae). Exp Appl Acarol 17:621–625
Kwong WK, Moran NA (2016) Gut microbial communities of social bees. Nat Rev Microbiol 14:374
Lee FJ, Rusch DB, Stewart FJ, Newton IL (2015) Saccharide breakdown and fermentation by the honey bee gut microbiome. Environ Microbiol 17:796–815
Ludvigsen J, Rangberg A, Avershina E, Sekelja M, Kreibich C, Amdam G, Rudi K (2015) Shifts in the midgut/pyloric microbiota composition within a honey bee apiary throughout a season. Microbes Environ 30:235–244
Luo QH, Zhou T, Dai PL, Song HL, Wu YY, Wang Q (2011a) Prevalence, intensity and associated factor analysis of Tropilaelaps mercedesae infesting Apis mellifera in China. Exp Appl Acarol 55:135–146
Luo Q, Zhou T, Wang Q, Dai PL, Wu YY, Song HL (2011b) Identification of tropilaelaps mites (Acari, Laelapidae) infesting Apis mellifera in China. Apidologie 42:485–498
Mohr KI, Tebbe CC (2006) Diversity and phylotype consistency of bacteria in the guts of three bee species (Apoidea) at an oilseed rape field. Environ Microbiol 8:258–272
Moran NA, Nakabachi A (2008) Genomics and evolution of heritable bacterial symbionts. Annu Rev Genet 42:165–190
Olofsson TC, Alsterfjord M, Nilson B, Butler E, Va’squez A (2014) Lactobacillus apinorum sp. nov., Lactobacillus mellifer sp. nov., Lactobacillus mellis sp. nov., Lactobacillus melliventris sp. nov., Lactobacillus kimbladii sp. nov., Lactobacillus helsingborgensis sp. nov. and Lactobacillus kullabergensis sp. nov., isolated from the honey stomach of the honeybee Apis mellifera. Int J Syst Evol Microbiol 64:3109–3119
Pakwan C, Kaltenpoth M, Weiss B, Chantawannakul P, Jun G, Disayathanoowat T (2017) Bacterial communities associated with the ectoparasitic mites Varroa destructor and Tropilaelaps mercedesae of honey bees (Apis mellifera). FEMS Microbiol. https://doi.org/10.1093/femsec/fix160
Powell JE, Martinson VG, Urban-Mead K, Moran NA (2014) Routes of acquisition of the gut microbiota of the honey bee Apis mellifera. Appl Environ Microbiol 80:7378–7387
Prakaimuk S, Li Y, Kanokporn S, Chen Z, Chantawannakul P (2015) Midgut bacterial communities in the giant Asian honeybee (Apis dorsata) across four developmental stages: a comparative study. Insect Sci 8:3–9
Pusceddu M (2016) Social immunity in honeybee: behavioral, chemical and microbiological aspects. Dipartimento di Agraria, Sezione di Patologia Vegetale ed Entomologia, Università degli Studi di Sassari, Viale Italia, Sassari
Quast C, Pruesse E, Yilmaz P, Gerken J, Schweer T, Yarza P, Peplies J, Glöckner FO (2013) The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res 41:D590–D596
Rosenkranz P, Aumeier P, Ziegelmann B (2010) Biology and control of Varroa destructor. J Invertebr Pathol 103:96–119
Sandionigi A, Vicario S, Prosdocimi EM, Galimberti A, Ferri E, Bruno A, Balech B, Mezzasalma V, Casiafhi M (2015) Towards a better understanding of Apis mellifera and Varroa destructor microbiomes: Introducing ‘phyloh’ as a novel phylogenetic diversity analysis tool. Mol Ecol Resour 15:697–710
Saraithong P, Li Y, Saenphet K, Chen Z, Chantawannakul P (2017) Midgut bacterial communities in the giant Asian honeybee (Apis dorsata) across 4 developmental stages: a comparative study. Insect Sci 24:81–92
Schneider DS, Ayres JS (2008) Two ways to survive infection: what resistance and tolerance can teach us about treating infectious diseases. Nat Rev Immunol 8:889–895
Snowdon JA, Cliver DO (1996) Microorganisms in honey. Int J Food Microbiol 31:1–26
Vanbergen AJ (2013) Threats to an ecosystem service: pressures on pollinators. Front Ecol Environ 11:251–259
Vanengelsdorp D, Meixner MD (2010) A historical review of managed honey bee populations in Europe and the United States and the factors that may affect them. Invertebr Pathol 103:S80–S95
Vásquez A, Forsgren E, Fries I, Paxton RJ, Flaberg E, Szekely L, Olofsson TC (2012) Symbionts as major modulators of insect health: lactic acid bacteria and honeybees. PLoS ONE 7:e33188
Wang Q, Garrity GM, Tiedje JM, Cole JR (2007) Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microbiol 73:5261–5267
White DC, Sutton SD, Ringelberg DB (1996) The genus Sphingomonas: physiology and ecology. Curr Opin Biotechnol 7:301
Yun JH, Jung MJ, Kim PS, Bae JW (2018) Social status shapes the bacterial and fungal gut communities of the honey bee. Sci Rep 8:2019
Zhang J, Zhang Y, Han R (2016) The high-throughput production of dsRNA against sacbrood virus for use in the honey bee Apis cerana (Hymenoptera: apidae). Virus Genes 52:698–705
Acknowledgements
We thank Pinhong Wang, Guirong Li and Yujuan Qiu (Institute of Apicultural Research, China Academy of Agricultural Sciences) for beekeeping. This work was supported by the Beijing Natural Science Foundation (No. 6162026), the Central Public-interest Scientific Institution Basal Research Fund (IAR-CPSIBRF-2017-1) and the Agricultural Science and Technology Innovation Program (CAAS-ASTIP-2017-IAR).
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Ma, S., Yang, Y., Jack, C.J. et al. Effects of Tropilaelaps mercedesae on midgut bacterial diversity of Apis mellifera. Exp Appl Acarol 79, 169–186 (2019). https://doi.org/10.1007/s10493-019-00424-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10493-019-00424-x